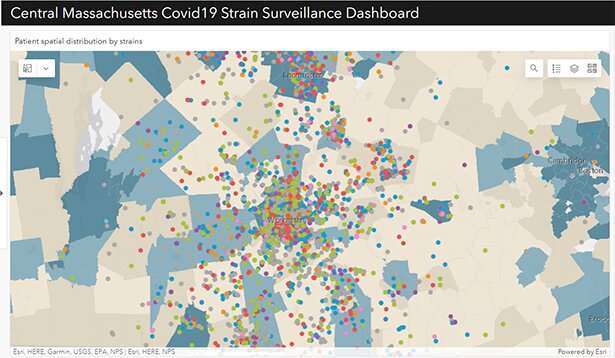
The dashboard allows epidemiologists and public health officials to access a complex and diverse set of data about the COVID-19 pandemic in Central Massachusetts in a user-friendly application. Credit: University of Massachusetts Medical School
Geneticist Doyle Ward, Ph.D., and colleagues at UMass Chan Medical School have developed a web app that allows epidemiologists, public health officials and other scientists to access genetic information about SARS-CoV-2 virus from Central Massachusetts. The study was published in JMIR Formative Research.
Created by Qiming Shi, Ph.D., a data scientist in the Center for Clinical and Translational Science at UMass Chan, the app, called MAGGI (University of Massachusetts' Graphical user interface for Geographic Information), draws on health-associated social demographic data and viral genomic data collected from more than 6,000 SARS-CoV-2 cases in Central Massachusetts. By linking temporal, spatial, demographic and de-identified clinical data to COVID-19 genetic variants, local epidemiologists and public health officials now have another tool to monitor and forecast the progress of the COVID-19 pandemic and potentially track the impact of interventions.
"The dashboard allows epidemiologists and public health officials to access a complicated and diverse set of data about the COVID-19 pandemic in Central Massachusetts in a user-friendly application," said Dr. Ward, associate professor of microbiology & physiological systems and operations director at the Center for Microbiome Research at UMass Chan. "With this tool, researchers can look at the data graphically in a meaningful way that lets them understand what the fundamental data is and how to apply it."
The data that forms the backbone of MAGGI was collected from a number of public sources, including local vaccination rates from the Massachusetts Department of Public Health and social vulnerability indicators from the Centers for Disease Control and Prevention. Additionally, zip code-level demographic data was sourced from the Zip Code Tabulation Areas and the 2018 American Community Survey.
Ward and colleagues worked with UMass Memorial Health to obtain and archive remanent SARS-CoV-2 positive samples collected in clinical settings. They used deep sequencing technology to reconstruct individual viral genomes and identify the variant of each COVID-19 sample.
Finally, de-identified clinical data, linked to each of the thousands of COVID-19 samples, was imported from the clinical data research warehouse maintained by UMass Chan. This data combines information from the electronic health systems at UMass Memorial but with all personal information that could be used to identify individual people removed.
The result is a map of Central Massachusetts with thousands of COVID-19 cases represented by colored dots coded for genetic sub-types and plotted across the region by zip code.
"We think this is unique to Central Massachusetts," Ward said. "Nobody else has COVID-19 information at this level of detail."
MAGGI is more than a simple map though; it is also overlaid with social and clinical data that researchers can use to spot patterns and respond to the pandemic. Users can access information about vaccine rates, social vulnerability, age, gender, poverty rates, the date the sample was taken, potential comorbidities, hospitalizations, clinical outcomes, and more on top of the genetic, geographic and temporal information.
With access to this data, public health officials can potentially observe how the pandemic is progressing almost in real time. As the shape of the pandemic evolves, health officials can potentially formulate new responses and interventions aimed at flattening infection rates or gear up for an influx of hospitalizations, for example. Additionally, scientists can study the data to gain new insights into the virus, such as how it impacts local populations.
"We can use MAGGI to scale the timing and location of COVID-19 infection and visualize associations and patterns between vaccine rates, and social vulnerability on a street level," said Carly Herbert, a doctoral student in the Morningside School of Biomedical Sciences and co-author on the study. "You can imagine how useful this data can be from a clinical perspective. We can use it to track emerging variants, assess interventions and identify if a variant is behaving differently, spreading more rapidly or causing more hospitalizations."
Ward said MAGGI and the complex data sets behind it will allow researchers to identify and observe patterns in near real-time and get to the root causes of those patterns. "It takes about a month between when we receive the sample to when we get a genetic sequence and the corresponding data into MAGGI. The hope is this information can be used to fashion a response to emerging trends or patterns," he said.
Because MAGGI is built on several publicly available data sets, it could potentially be replicated in other locations as well. To get the same level of specificity would require access to high throughput sequencing technologies as well as de-identified clinical data.
Beth A. McCormick, the Worcester Foundation for Biomedical Research Chair, professor and vice chair of microbiology & physiological systems and founding director of the Center for Microbiome Research, said underneath all this data is a unique collaboration between UMass Chan and UMass Memorial.
"Because researchers at UMass Chan work so closely with our clinical partners at UMass Memorial and they are the primary provider of health care for the Central Massachusetts region, we have a unique opportunity to collaborate on public health projects such as MAGGI," said Dr. McCormick. "The real strength of this study, in our minds, is how we were able to get such an amazing level of detail while also protecting patient privacy. That doesn't happen without close collaboration between scientists and health providers."
Ward and colleagues continue to sequence new viral samples for the project as they arrive from the clinic. The hope, as collection protocols are improved and streamlined, is to improve the turn around on sequencing to a few weeks or days to better inform public health decisions.
More information: Qiming Shi et al, COVID-19 Variant Surveillance and Social Determinants in Central Massachusetts: Development Study, JMIR Formative Research (2022). DOI: 10.2196/37858
Provided by University of Massachusetts Medical School