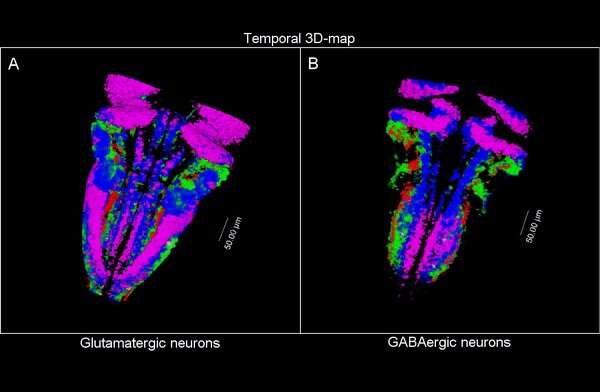
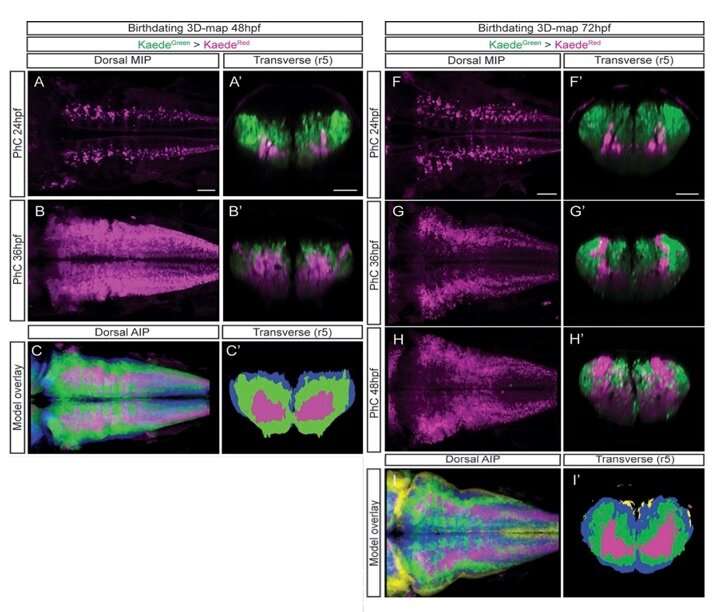
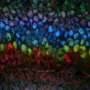
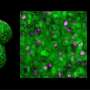
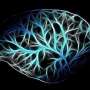
3D time map of different neuronal populations. Credit: Universitat Pompeu Fabra - Barcelona
The tissues of every living organism develop from a few cells that divide and move until they reach their final destination and position. The process of how a certain tissue or organ is formed is called morphogenesis, and the birth of neurons is known as neurogenesis.
Researchers from the UPF Neurodevelopmental Dynamics Research Group, led by ICREA Acadèmia professor and researcher Cristina Pujades, have studied these two processes using the hindbrain of the zebrafish. They have thus created a 3D atlas that allows for finding out when neuronal differentiation in its definitive form occurs and, moreover, they have shown that the neurons' final position in the tissue depends on their time and place of birth. The results of the research have been recently published in the journal eLife.
"The microscopy and image analysis techniques we currently dispose of are impressive and allow us to see in real time the movement of cells and the formation of many biological structures," Pujadas explains. "But quantifying the biology that we observe is very complicated, in general in biology it is increasingly difficult to quantify and measure. For example, quantifying the volume of the brain is very complex since it is not homogeneous, it is not like a football that has the same uniformly distributed layers."
Currently there are tools that allow quantifying in 2D, but very few do so in 3D. In collaboration with the European Molecular Biology Laboratory of Barcelona (EMBL Barcelona), the team developed a pipeline, or a program that contains various tasks or functions, to generate 3D models from microscopic images combining different signals that allow viewing the internal structures. To achieve this they used DAMAKER, a platform they themselves developed.
Transition from 2D to 3D. Tracking temporal changes in neuronal growth during brain morphogenesis. Credit: Universitat Pompeu Fabra - Barcelona
So, they combined algorithms already published for other functions and coupled them differently to be able to apply them to answer their biological questions: how do neuron populations grow? And how are excitatory (glutameric) and inhibitory (GABAergic) neurons generated?
"We saw that by using fluorescent markings that allowed us to visualize a specific cell population, we could use them as biomarkers throughout the life history of each cell during tissue formation and also to calculate the volume they occupy. This allows us to visualize and quantify parameters that we couldn't previously see."
The researchers have created a 3D brain atlas from confocal microscopy images and computational tools to allow understanding when neurons are born and what their final destination is. But, above all, "this allows us to infer, from their position in the brain, what their date of birth was, since there is an internal-external gradient. This means that the neurons that were differentiated before are in the innermost part of those that were born later, which allows us to study the dynamics of the different neural populations," Pujades says.
The part of the hindbrain that they have studied (the rhomboencephalic) is highly conserved in vertebrates, that is, that there is no difference between humans and lampreys. It is also known as the primitive hindbrain because that is where the basic functions of vertebrates are controlled, such as swallowing, breathing, innervation of the viscera, etc.
What is the advantage of this? For example, human avatars can be created in zebrafish, that is, we can generate the mutation we need in fish. "We observe how it develops and instead of looking for the neural affections as if it were a haystack, we can go to a certain status, look at the distribution of the neurons and, by comparing it to the atlas, we can know when the neural defect took place," Pujadas says.
This is the first mapping of the brain carried out in a vertebrate, since cartographies of the fruit fly already existed. The system is written in open source and can be applied to any organ, for example the heart, and can also be used in any model, for example fly or mouse. It only requires the specific markers of what the researcher wants to see.
More information: Matthias Blanc et al, A dynamic and expandable Digital 3D-Atlas MAKER for monitoring the temporal changes in tissue growth during hindbrain morphogenesis, eLife (2022). DOI: 10.7554/eLife.78300
Journal information: eLife
Provided by Universitat Pompeu Fabra - Barcelona