Artificial intelligence methods may replace histochemical staining
Pathologists observe tissue samples by staining them first. However, the standard procedures for staining tissue samples in histopathology are time-consuming and require specialized laboratory infrastructure, chemical reagents, and skilled technicians. Uncertainty in tissue staining in the handling of different laboratories and histology technicians may lead to misdiagnosis. In addition, the original tissue sample is not preserved by these histochemical staining techniques currently in use since each step of the procedures has irreversible impact on the sample.
With the advancement of artificial intelligence (AI), researchers are using AI techniques to improve pathology workflow. A recent study from the University of California Los Angeles (UCLA) used deep neural networks to virtually stain microscopic images of unlabeled tissue. The research was published in Intelligent Computing.
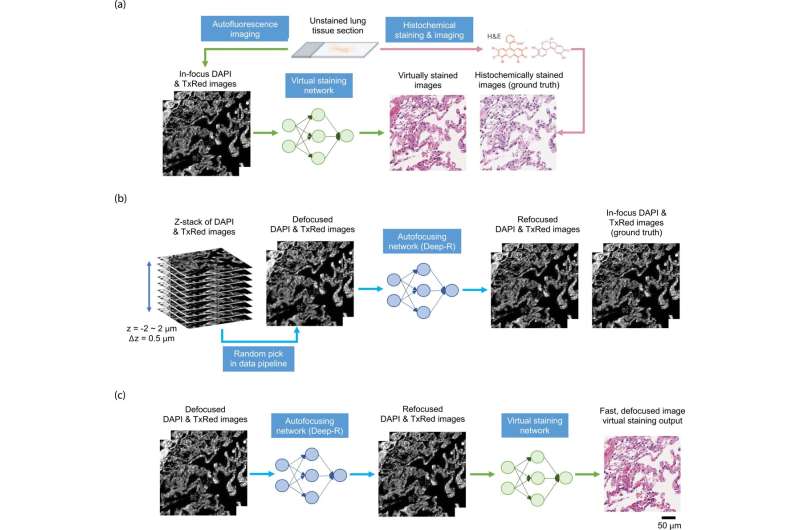
Deep neural networks have already been applied to stain unlabeled tissue section images, avoiding different laborious and time-consuming histochemical staining processes. There are, however, some bottlenecks. "In all the label-free virtual staining methods, the acquisition of in-focus images of the unlabeled tissue sections is essential. In general, focusing is a critical but time-consuming step in scanning optical microscopy," the authors said.
The most widely used autofocusing method demands many focus points across the tissue slide area with high focusing precision, and the best focal plane is determined by an iterative search algorithm, which is time consuming and may introduce photodamage and photobleaching on the samples.
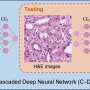
To overcome these problems, the authors present a new deep learning-based fast virtual staining framework. They say that "this framework uses an autofocusing neural network (termed Deep-R) to digitally refocus the defocused autofluorescence images. Then a virtual staining network is used to transform the refocused images into virtually stained images."
Compared to the standard virtual staining framework, the new framework demonstrated by the authors uses fewer focal points and reduces the focusing precision for each focus point to acquire coarsely-focused whole slide autofluorescence images of tissue.
This new virtual staining framework can significantly reduce the time for autofocusing and the entire image acquisition process. The authors say that "the deep learning-based framework decreases the total image acquisition time needed for virtual staining of a label-free whole slide images (WSI) by ~32%, also resulting in a ~89% decrease in the autofocusing time per tissue slide."
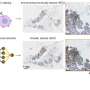
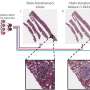
Despite loss of image sharpness and contrast compared to standard virtual staining frameworks, high quality staining can still be produced, closely matching the corresponding histochemically stained ground truth images. Furthermore, this framework can also be used as an add-on module to improve the robustness of the standard virtual staining framework.
This fast virtual staining framework will have more development prospects in the future. "This fast virtual staining workflow can also be expanded to many other stains, such as Masson's Trichrome stain, Jones' silver stain, and immunohistochemical (IHC) stains," the authors said. "Although the virtual staining approach presented here was demonstrated based on the autofluorescence imaging of unlabeled tissue sections, it can also be used to speed up the virtual staining workflow of other label-free microscopy modalities."
More information: Yijie Zhang et al, Virtual Staining of Defocused Autofluorescence Images of Unlabeled Tissue Using Deep Neural Networks, Intelligent Computing (2022). DOI: 10.34133/2022/9818965