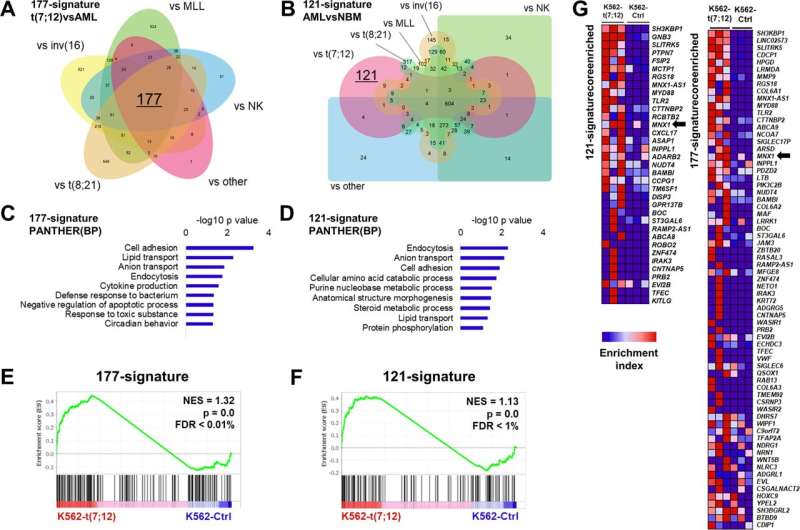
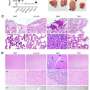
Comparison of K562-t(7;12) transcriptional landscape with t(7;12) patient signatures. A 177-gene signature of t(7;12)-patient gene expression extrapolated from published microarray and RNA sequencing datasets by comparison with other pediatric AML subtypes. The Venn diagram shows the 177 intersect genes. B 121-gene t(7;12)-specific signature inferred by comparisons of gene expressions of pediatric AML and normal bone marrow (NBM) samples. Edward’s Venn diagram highlight the 121 exclusive genes to t(7;12). C, D Gene Ontology analysis of the 177-signature and 121-signature by the PANTHER annotation repository of biological processes (BP). E, F GSEA enrichment plots of K562-t(7;12) gene expression profile using the 177- and 121-signatures. NES normalized enrichment score, FDR false discovery rate. G Core enriched genes from the GSEA using the 177- and 121-signatures, shown by their enrichment index in K562-t(7;12) against K562-Ctrl. MNX1 is highlighted by the arrows. Credit: Oncogenesis (2022). DOI: 10.1038/s41389-022-00426-2
Babies with leukemia could get an array of new treatments after scientists used genetic engineering to reproduce a gene defect found in the disease.
Acute myeloid leukemia is a high-risk cancer which affects children under two.
Scientists first pinpointed a biomarker linked to the cancer about twenty years ago.
Now for the first time they've used genetic engineering to make a model of the glitch which means they can study its biological mechanisms to unravel potential drug targets.
"This is the first step towards finding a cure for this rare but deadly form of childhood leukemia," said Dr. Sabrina Tosi at Brunel University London.
"By generating an in vitro model for this type of leukemia, we are providing a tool for further investigation. It has enabled us to identify potentially druggable targets leading to possible new treatment. "
Every year in the U.K. about 100 children are diagnosed with acute myeloid leukemia. And there's no single "go to" treatment—doctors try different combinations of chemotherapy and bone marrow (stem cell) transplantation. Nearly no young children survive longer than three years.
Too much of the gene MNX1 product is found in infants with acute myeloid leukemia carrying a particular gene alteration Dr. Tosi discovered in 2000. But because the disease mainly affects babies, it is difficult to collect enough cancerous cells from the patients to study, so researchers know little about how the gene works. Her team has used gene editing to make a cellular model containing the genetic alteration that over-produces MNX1 that can be recreated over and over again. This means scientists can now study it intensely to see how it works at a molecular level, which will help highlight new treatments.
"Thanks to recent advances in genome engineering technologies, novel murine and human models have been developed," says the study published in the journal Oncogenesis. "Mechanisms of leukemogenesis are therefore beginning to be uncovered, including the exact developmental window affected and the role of MNX1. Understanding the cytogenetic, molecular, and clinical features of t(7;12) will pave the way towards targeted therapeutic interventions."
"In children, the standard therapy right now is essentially the same as for adults. Chemotherapy and, in some cases, stem cell transplantation," explained Doctoral Researcher Denise Ragusa. "So, the hope would be that we could identify something that is specific to this particular type of leukemia so therapies are more effective and not as aggressive."
More information: Denise Ragusa et al, Engineered model of t(7;12)(q36;p13) AML recapitulates patient-specific features and gene expression profiles, Oncogenesis (2022). DOI: 10.1038/s41389-022-00426-2
Journal information: Oncogenesis
Provided by Brunel University