New proteomic technique reveals secrets of dendrite development
NeuroOmics technology lets researchers label and capture cell-surface proteins in intact, live tissue—opening opportunities to understand complex cellular interactions and future drug targets.
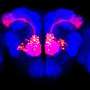
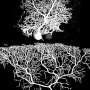
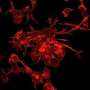
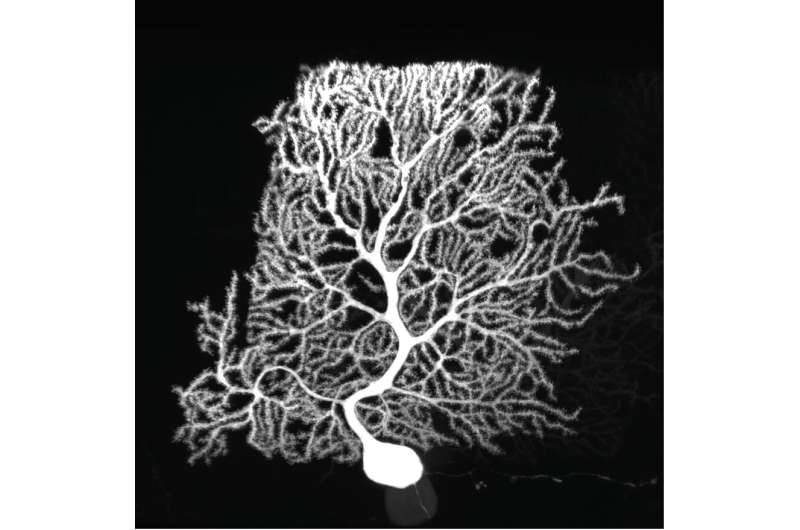
Purkinje cells are large neurons that form in the cerebellum, the small compact portion of the brain behind the cerebrum. Intricate, dendritic trees extend from each cell, forming a convoluted branching pattern that resembles a sea fan. Dysfunction of Purkinje cells has been linked to neurological symptoms, such as tremors, irregular muscle movement and hyperreactivity.
Purkinje cells' elaborate dendrites play an integral role in neuronal communication, by receiving and integrating synaptic impulses that are coordinated by proteins scattered across the surface of the cell. Despite this integral role in neural computation, an understanding of the development of these elaborate dendrites remains elusive. A study published online in the October issue of Neuron applies a new technology to profile the proteins present on the surface of Purkinje cells to gain a better understanding of their role in dendrite development and neuronal communication.
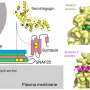
Cell-surface proteins stick out of the cell membrane, like antennae on a building. By protruding into the surrounding environment, the proteins play a critical role in intercellular communication, like the highly orchestrated interactions between the different cell types in the nervous systems of mammals. Most techniques for studying these proteins are unable to quantify and profile the proteins in the diverse cell-type mosaic of the mammalian brain.
"Cell surface proteins are like little machines on the outside surface of cells that mediate communication between different types of cells," said study lead author Andrew Shuster, a postdoc at Harvard University and former member of the Luo lab. "We were interested in developing a method to profile the entire coat of cell surface proteins in healthy tissue that has not been approachable by other methods."
The new technology, called in situ cell-surface protein extraction by extracellular labeling (iPEEL), enables precise labeling of surface proteins that extend from the cell surface. iPEEL is a product of Wu Tsai Neurosciences Institute's Neuro-omics Initiative, which aims to develop tools to map gene and protein expression in the nervous system as well as neuronal connection patterns.
"We started this endeavor together with a few labs at Stanford by borrowing and extending technology developed for cell cultures," said study senior author Liqun Luo, the Ann and Bill Swindells Professor at Stanford University and a Howard Hughes Medical Institute Investigator. "This approach, extended from our collaborator Alice Ting's work in cultured cells, will be useful for a whole slew of organ systems and many different applications to study cell development and disease."
To date, the full complement of cell surface proteins had not been described for any cell type in healthy mammalian tissue. Luo and his team applied the technology to Purkinje cells obtained from transgenic mouse tissues. Using iPEEL, they labeled and profiled the proteins that line the surface of developing (15 days old) and mature (35 days old) Purkinje cells. The aim was to determine how the composition of surface proteins changes with age and to explore their function.
They identified a subset of proteins enriched in young Purkinje cells that might be involved in dendrite growth and branching. Further work assessing the functions of these proteins brought them to the Armadillo-like helical domain-contain protein 4 (Armh4), a protein with no known function in the nervous system.
They found that Armh4 actually plays a critical role in the process by which developing neurons produce their elaborate dendrites. By disrupting the Armh4 protein by either loss-of-function or overexpression, the researchers were able to impair the shape and development of the dendritic branches. Some of their findings also suggest that Armh4 impacts synapse formation and may be a link between synaptogenesis and dendrite morphology.
"Cell surface proteins have important functions for many cell types that biologists will find exciting," said Shuster. "[Armh4] is expressed in many cell types across the nervous system. Studying this protein will elucidate its role in regulatory functions and provide an opportunity to understand the pathways it regulates."
Their work also revealed that many of the same synaptic and channel proteins were active in both developing and mature cells, suggesting that active engagement of electrical and synaptic signaling was possible even before mature neural circuitry had been established. This finding expands on the understanding of how the neuronal surface evolves as the cell matures.
Beyond identifying the full complement of cell surface proteins for mammalian tissues, this technology provides a new platform to explore drug development. Today many drugs target these proteins to treat disease. iPEEL opens opportunities to identify new drug targets.
"We can map the entire cell surface proteome for a variety of cells," said Luo. "This technology will allow researchers to study different animal models to explore therapeutic pathways to treat disease and find new drug targets that can aid diseased tissues."
More information: S. Andrew Shuster et al, In situ cell-type-specific cell-surface proteomic profiling in mice, Neuron (2022). DOI: 10.1016/j.neuron.2022.09.025