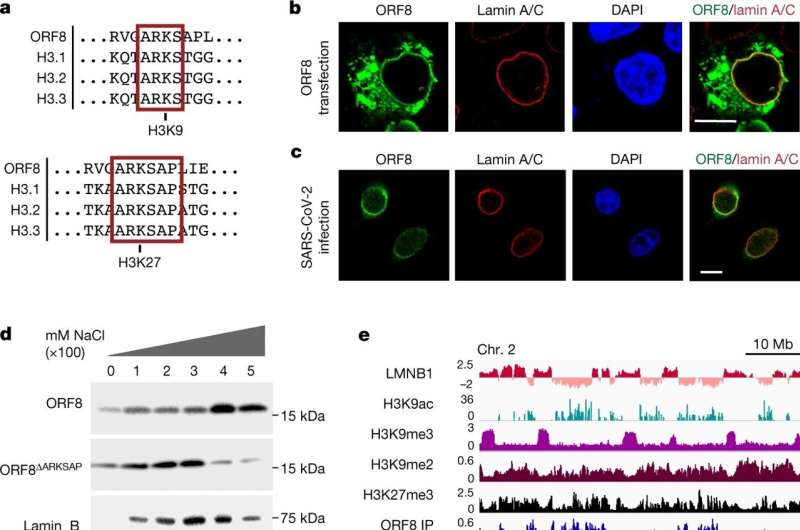
ORF8 associates with chromatin. a, ORF8 contains an ARKS motif at amino acid 50 that matches the histone H3 tail. b, Lamin A/C staining of HEK293T cells transfected to express Strep–ORF8. c, ORF8 and lamin A/C staining of SARS-CoV-2-infected A549ACE2 cells at MOI = 1, 48 h after infection. d, Sequential salt extraction of HEK293T cells expressing ORF8 or ORF8ΔARKSAP. e, Gene tracks for ORF8 ChIP–seq normalized to input controls. f, Targeted mass spectrometry analysis of trypsin-digested ORF8 showing that ORF8 is acetylated at lysine 52. The intact peptide or precursor at 879.9508 m/z with a 2+ charge was isolated and fragmented. Tandem mass spectrometry spectra show unfragmented precursor (green) with matching product ions within a mass error of 10 ppm. Fragment intensity is relative to that for the ion with the highest intensity across the m/z range. The color, letter and number for each fragment indicate the sequence that fragment contains within the larger peptide (top). y (red) and b (blue) fragments indicate C- and N-terminus-matched fragments, respectively. g, ORF8 expression results in decreased levels of KAT2A. Scale bars, 10 μm. Credit: Nature (2022). DOI: 10.1038/s41586-022-05282-z
A team of researchers at the University of Pennsylvania, working with a colleague from the University of Texas Medical Branch and another from Boston University and Boston Medical Center, has found that the SARS-CoV-2 virus produces a protein that mimics a protein that packages DNA, preventing transcription that would normally play a role in an immune response.
In their paper published in Nature, the group describes how they compared proteins that package human DNA with proteins produced by the SARS-CoV-2 virus and what doing so showed them. Lisa Thomann and Volker Thiel with the Institute of Virology and Immunology, Bern and Mittelhäusern, have published a News & Views piece in the same journal issue describing the work done by the team on this new effort.
As the pandemic has worn on and lessened, medical researchers have continued to study the virus behind it, and one of the areas they have focused on is its remarkable ability to suppress a response by host cells in those infected. In this new effort, the researchers have taken a closer look at the relationships between proteins that package DNA in human cells and proteins produced by the SARS-CoV-2 virus.
Prior research has shown that the SARS-CoV-2 virus produces three main types of proteins; those involved in replication, those involved in structural processes and a third that are known as accessory proteins.
The third type is involved in a wide range of activities. Prior research has also shown that human DNA is packaged in proteins known as histones—they serve to hold the DNA strands in place and to serve as gatekeepers for materials that read the strands and use that information to engage in activities such as signaling to other body parts. Some such signaling is involved when a host is infected to urge defense mechanisms to fight the intrusive agent.
In this new effort, the researchers have found evidence that suggests that one of the accessory proteins produced by the SARS-CoV-2 virus (ORF8) is able to mimic at least one type of histone protein (KAT2A) that packages DNA. And by doing so dampens the type of signaling that is supposed to occur when a host is infected, reducing the immune response.
More information: John Kee et al, SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry, Nature (2022). DOI: 10.1038/s41586-022-05282-z
Lisa Thomann et al, SARS-CoV-2 mimics a host protein to bypass defences, Nature (2022). DOI: 10.1038/d41586-022-02930-2
Journal information: Nature
© 2022 Science X Network