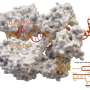
Generation and screening of a pooled library of CARs with diverse signaling domains. (A) Our CARs combine an αCD19 ScFv (FMC63), a CD8 hinge and transmembrane domain (TMD), an intracellular signaling domain, and a CD3ζ domain (left). Forty domains from across the human proteome were codon-optimized, synthesized, pooled into a plasmid library, and packaged to generate lentivirus (right). These spanned protein families such as immunoglobulin superfamily (IgSF); T cell/transmembrane, immunoglobulin, and mucin (TIM); TNF receptor superfamily (TNFRSF); and triggering receptors expressed on myeloid cells (TREM). (B) Venn diagram showing natural expression of library members across immune cell types. Expression patterns are listed in table S1. NKT, natural killer T cell; DC, dendritic cell. (C) Primary human CD4 and CD8 T cells were separately isolated from PBMCs for two human donors and lentivirally transduced with the library. The cells were FACS-purified using T2A-GFP fluorescence within one log of mean expression to reduce variability. (D) The pooled library was repeatedly stimulated 1:1 with CD19+ or CD19− irradiated K562 tumor cells to quantify antigen-specific activation (CD69), cytokine production (IFN-γ and IL-2), and proliferation [CellTrace Violet (CTV)]. The library was also sequenced at the specified time points to measure the relative expansion of individual constructs. (E) Percentage of CD8+ T cells expressing different numbers of exhaustion markers (PD1, TIM3, LAG3, and CD39) after a repeat stimulation assay with CD19+ irradiated K562 tumor cells. T cells expressing CD28, 4-1BB, or CD3ζ-only CARs are compared to untransduced (Untrans.) cells. Gray boxes correspond to time points in which no live cells remained. Significance was assessed in CD8 T cells for two donors using a repeated-measures ANOVA model. FDR-corrected *P = less than 0.05 and ***P = less than 0.001; ns, not significant. The second donor and data for CD4+ T cells are shown in fig. S1 (A and B). (F) We used FlowSeq, a FACS and next-generation sequencing-based pooled quantification workflow, to quantify enrichment by sorting the library into bins of fluorescent signal corresponding to a functional readout, as shown by the colored histograms. We then amplified the costimulatory domain by genomic DNA extraction and PCR and performed amplicon sequencing on each bin to estimate the phenotype for each library member. Credit: Science Translational Medicine (2022). DOI: 10.1126/scitranslmed.abm1463
In recent years, genetically re-engineered immune cells—armed with molecular weaponry to recognize and destroy tumor cells—have changed the landscape of cancer treatment. Now, UC San Francisco researchers have developed a new method for comparing massive numbers of these CAR-T cells, each with slightly different molecular features, to determine which is most effective and long-lasting against cancer.
In their study published Nov. 9, 2022, in Science Translational Medicine, the team used the approach—dubbed "CAR Pooling"—to study CAR-T cells with forty different receptors. The screen, which can be expanded to test hundreds or thousands more receptor combinations in the future, has already revealed new and surprising receptors that make these therapeutic cells more powerful.
"CAR-T cells have been absolutely transformative for a lot of people with blood cancers," said senior author Kole Roybal, Ph.D., UC San Francisco associate professor of microbiology and immunology and a member of the Gladstone-UCSF Institute of Genomic Immunology. "This work is a stepping stone toward engineering these cells in even smarter ways so they work better, for longer, and in more cancer types."
A transformative treatment
T cells are a type of white blood cell expressing receptors on their surface that recognize foreign material in the body. When a matching molecule or particle fits into the T cell receptor, the cell launches an immune response to ward off the invader. This kind of response can destroy not only viruses and bacteria, but cancer cells as well.
When a cancer patient is being treated with CAR-T cell therapy, clinicians collect T cells from the patient's blood or from a healthy donor. Then, they alter these cells in the lab, adding DNA that coaxes the immune cells into producing special, cancer-recognizing chimeric antigen receptors (CARs) on their surface. The modified cells are grown and injected back into the patient. The new CARs help the T cells specifically attack cancer cells. A variety of CAR-T cells have been approved for use in blood cancers, including lymphomas, leukemias and multiple myeloma.
"Our ability to take T cells out of the body, add new genetic programs to them, and put them back into the body as a living programmable therapy has already made a huge impact on the treatment of hematological cancers," said Daniel Goodman, Ph.D., a UCSF postdoctoral scholar and co-first author of the new study. "However, this is only the beginning of what engineered cells could be capable of if we understood more fully how to manipulate their signaling and functional properties."
Researchers have struggled to get CAR-T cells working against solid tumors. Even for many patients with blood cancer, CAR-T cells work only temporarily or not at all.
One of the limiting factors in developing new CAR-T cell therapies is the high-throughput technology required to modify and test new receptors; across the immune system there are hundreds of receptor pieces which can fit together like Lego bricks into thousands of possible combinations.
Streamlining T cell tests
Roybal, Goodman and co-first author Camillia Azimi developed CAR Pooling to rapidly test the effectiveness of different CARs at once. The screen works by generating a DNA library containing the genetic codes for many immune receptors, simultaneously introducing them to millions of cells, and then having the cells compete—in test tubes—for their skills in surviving, growing and recognizing and destroying cancer.
"Our method makes the process of testing new receptor based-therapeutics much faster," says Azimi, a UCSF graduate student at the time. "This not only allows the researchers to save time, but also to explore designs they wouldn't have been able to test individually."
Among the 40 CARs tested in the new study, for instance, were not only classic T-cell receptors but receptors borrowed from other types of related immune cells. For example, a B-cell receptor named BAFF-R is the leading contender for making a T cell particularly effective against cancer.
"We would never have chosen to test BAFF-R if we weren't utilizing CAR Pooling," says Azimi.
When the CAR-T cell containing BAFF-R was tested in mice with multiple myeloma, the mice survived longer and had less cancer compared to mice that received standard CAR-T cells.
A platform for future studies
The research team is planning to expand the use of CAR Pooling to test even more receptors in re-engineered T cells. Eventually, Roybal said, the approach could be used to develop different varieties of CAR-T cells for different types of cancer; there's likely no one-size-fits-all answer when it comes to diverse cancer types.
"We could certainly use CAR Pooling to rapidly find the best receptors for specific clinical settings," he says. "One set of receptors may be more beneficial for glioblastoma but another most beneficial for myeloma."
As the researchers screen more and more immune receptors, they hope to reveal underlying patterns in how the receptors dictate the activity of immune cells. Eventually, this could lead to even more synthetic receptors that can take on not only cancer, but other diseases.
"We hope to learn how to compose the fundamental pieces of these signaling domains and build the next generation of engineered cell therapies," says Goodman.
The lab is also working on understanding and improving other aspects of CAR-T cells, beyond their receptors, to boost the effectiveness of the therapy.
"There are a lot of ways you can modify these cells," says Roybal. "This is just one piece of the puzzle."
More information: Daniel B. Goodman et al, Pooled screening of CAR T cells identifies diverse immune signaling domains for next-generation immunotherapies, Science Translational Medicine (2022). DOI: 10.1126/scitranslmed.abm1463
Journal information: Science Translational Medicine
Provided by University of California, San Francisco
![Generation and screening of a pooled library of CARs with diverse signaling domains. (A) Our CARs combine an αCD19 ScFv (FMC63), a CD8 hinge and transmembrane domain (TMD), an intracellular signaling domain, and a CD3ζ domain (left). Forty domains from across the human proteome were codon-optimized, synthesized, pooled into a plasmid library, and packaged to generate lentivirus (right). These spanned protein families such as immunoglobulin superfamily (IgSF); T cell/transmembrane, immunoglobulin, and mucin (TIM); TNF receptor superfamily (TNFRSF); and triggering receptors expressed on myeloid cells (TREM). (B) Venn diagram showing natural expression of library members across immune cell types. Expression patterns are listed in table S1. NKT, natural killer T cell; DC, dendritic cell. (C) Primary human CD4 and CD8 T cells were separately isolated from PBMCs for two human donors and lentivirally transduced with the library. The cells were FACS-purified using T2A-GFP fluorescence within one log of mean expression to reduce variability. (D) The pooled library was repeatedly stimulated 1:1 with CD19+ or CD19− irradiated K562 tumor cells to quantify antigen-specific activation (CD69), cytokine production (IFN-γ and IL-2), and proliferation [CellTrace Violet (CTV)]. The library was also sequenced at the specified time points to measure the relative expansion of individual constructs. (E) Percentage of CD8+ T cells expressing different numbers of exhaustion markers (PD1, TIM3, LAG3, and CD39) after a repeat stimulation assay with CD19+ irradiated K562 tumor cells. T cells expressing CD28, 4-1BB, or CD3ζ-only CARs are compared to untransduced (Untrans.) cells. Gray boxes correspond to time points in which no live cells remained. Significance was assessed in CD8 T cells for two donors using a repeated-measures ANOVA model. FDR-corrected *P = less than 0.05 and ***P = less than 0.001; ns, not significant. The second donor and data for CD4+ T cells are shown in fig. S1 (A and B). (F) We used FlowSeq, a FACS and next-generation sequencing-based pooled quantification workflow, to quantify enrichment by sorting the library into bins of fluorescent signal corresponding to a functional readout, as shown by the colored histograms. We then amplified the costimulatory domain by genomic DNA extraction and PCR and performed amplicon sequencing on each bin to estimate the phenotype for each library member. Credit: Science Translational Medicine (2022). DOI: 10.1126/scitranslmed.abm1463 'CAR pooling' screens identify most effective cancer immunotherapy cells](https://scx1.b-cdn.net/csz/news/800a/2022/car-pooling-screens-id.jpg)