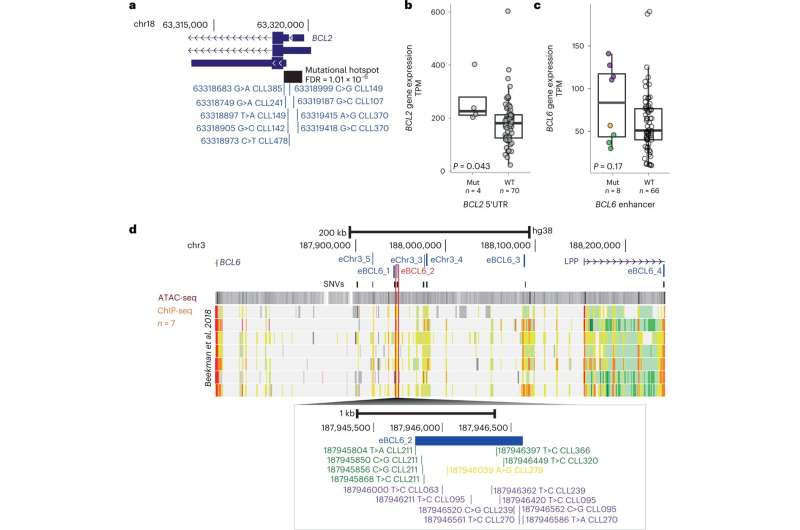
Noncoding mutations impacting BCL genes. a, Genome view of BCL2 5′ UTR. The significantly mutated region is indicated by a black rectangle. Individual somatic mutations are shown in blue. b, Gene expression of BCL2 in TPM determined by RNA-seq in samples with BCL2 5′ UTR mutations versus WT. Black dots are marks as outliers. P value was derived from a two-sided Welch’s t-test. c, Gene expression in TPM determined by RNA-seq of BCL6 in samples with BCL6 enhancer mutations versus WT. Expression levels were split in low (less than median expression; green), medium (between median expression and 100; orange) and high (≥100 TPM; purple). P value was derived from a two-sided Wilcoxon test. d, Genome view of the BCL6 gene and enhancers. Enhancers within these regions are annotated in blue font. eBCL6_2, which was the target of several variants is indicated in red. Annotation tracks of ATAC-seq and ChIP–seq are from publicly available RE annotation detailed above (references containing detailed of datasets and figure legends). The lower panel shows the individual mutations color coded as defined in c. All boxplots show the minimum and maximum values and interquartile range. Credit: Nature Genetics (2022). DOI: 10.1038/s41588-022-01211-y
A collaborative study led by the University of Oxford as part of the UK's 100,000 Genomes Project, published in Nature Genetics today, has defined five new subgroups of the most common type of blood cancer, chronic lymphocytic leukemia (CLL) and associated these with clinical outcomes. This new method for risk stratifying patients could lead to more personalized patient care.
This is the first study to analyze all the relevant changes in DNA across the entire cancer genome, rather than targeted regions, to classify patients with cancer and link these subgroups to clinical outcomes.
Professor Anna Schuh, Department of Oncology, University of Oxford, who led the study, said, "We know that cancer is fundamentally a disease caused by changes in DNA that are acquired over the lifetime of an individual. The lab tools we currently use to predict whether or not a patient is likely to respond to a given therapy usually focus on single abnormalities in the cancer DNA and do not accurately predict the patient's clinical outcome. This is why we asked the simple question: can we increase the precision of current testing by looking at all the acquired DNA changes in cancer at once?"
This study analyzed the entire genome sequences from 485 patients with CLL who were enrolled in national clinical trials led by the Universities of Liverpool and Leeds, provided samples for the UK CLL Biobank in Liverpool and consented for their samples to be used in the 100,000 Genomes Project run by Genomics England.
By comparing the whole genome sequencing data from the cancer and healthy tissues in these patients, the team were able to map known and newly identified DNA changes, structural alterations, cancer mutational signatures and other global measures associated with CLL throughout the genome. They identified 186 distinct and recurrent genomic alterations and used these to define five genomic subgroups of CLL that associate with different clinical outcomes.
Following additional validation in patients receiving a wider range of treatments than those included in this study, particularly targeted therapies, these new genomic subgroups for CLL could be used to better guide the selection of therapies for improved patient outcomes.
This study paves the way for routine clinical application of whole genome analyses for risk stratification in other cancer types.
The team's analysis also identified 148 new putative genetic drivers of CLL. Future research on these drivers may uncover new mechanisms in CLL initiation and progression, with potential for the development of novel therapeutics.
Professor Sir Mark Caulfield, Vice Principal for Health for Queen Mary University of London and former Chief Scientist and lead of the 100,000 Genomes Project at Genomics England said, "This is the largest international genomic analysis of this blood cancer, which excitingly demonstrates the real value of whole genome sequencing from the 100,000 Genomes Project. It also harnesses the high-quality blood cancer samples from the Liverpool Chronic Lymphocytic Leukemia Biobank and associated clinical data collected by the Clinical Trial Units at Liverpool and Leeds universities as part of multi-center National Cancer Research Institute-supported clinical trials.
"Our work shows that the entire genome is superior in classifying patients into groups compared to the conventional targeted approaches and that we can predict response to treatment more precisely and in more patients."
More information: Pauline Robbe et al, Whole-genome sequencing of chronic lymphocytic leukemia identifies subgroups with distinct biological and clinical features, Nature Genetics (2022). DOI: 10.1038/s41588-022-01211-y
Journal information: Nature Genetics
Provided by Queen Mary, University of London