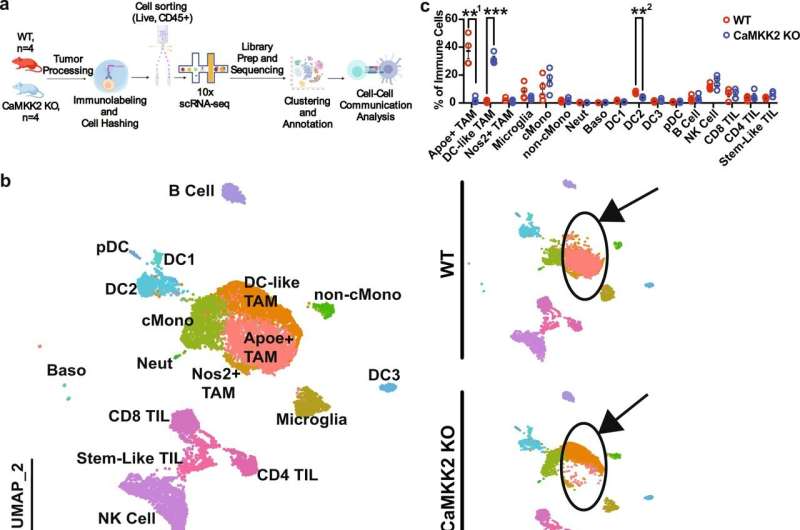
Pro-tumor immune programming in the glioma microenvironment is CaMKK2 dependent. a Schematic depicting scRNA-seq experimental design. n = 4 WT and CaMKK2 KO mice had 50k CT2a orthotopically implanted. On D14 tumors were harvested and underwent tumor processing. Samples were labeled with a Live-Dead viability Dye, CD45, and TotalSeqB anti-mouse Hashtag antibodies. CD45+ Live cells were sorted, and then pooled in equal parts by genotype. Graphic was created with BioRender.com. b 14k CD45+ Live Immune cells were sorted from WT and CaMKK2 KO tumor-bearing hemispheres on D14 p.i. HTO and Gene Expression libraries were prepared using the 10X platform. UMAP plots of the cell types identified using unsupervised clustering methods are shown for the aggregate dataset and stratified by genotype. c Abundance of immune cell types identified by scRNA-seq. n = 4 per genotype, two-way RM ANOVA p < 0.05 with post hoc unadjusted two-tail Fisher LSD t-test. **1p = 0.0059, ***p = 0.0003, **2p = 0.0016. Data are presented as mean ± SEM. d Dot plots corresponding to the cell types displayed in the UMAP plots show expression of subset-specific genes, with the dot size representing the percentage of cells expressing the gene and the color representing its average expression within a cluster. Credit: Nature Communications (2022). DOI: 10.1038/s41467-022-34175-y
Duke Health researchers have identified a unique process within the environment of deadly brain tumors that drives resistance to immune-boosting therapies and could be targeted to promote the effects of those drugs.
The finding, published in Nature Communications, explains a vexing problem in glioblastoma, a deadly form of brain tumor that has proven notoriously impervious to immune checkpoint inhibitors, a type of immunotherapy which has been highly effective in other cancers.
"Researchers have traditionally approached immunotherapy and resistance either by looking at cancer cells to find targets to attack, or by exploring the tumor environment for potential ways to improve the immune response," said lead author William H. Tomaszewski, Ph.D., who conducted the studies as a doctoral candidate in Duke's Department of Neurosurgery. "Our approach in this work was to find environmental factors that could make glioblastoma more vulnerable to immuno-therapies."
Tomaszewski and colleagues—including senior author John Sampson, M.D., professor of neurosurgery—focused on a molecule known as calmodulin-dependent kinase kinase 2, or CaMKK2, which is present in neurons and immune cells. This molecule is highly active in cancer supporting cells in the glioblastoma environment and is associated with worse survival in humans.
In mouse studies, the cancer-killing T-cells were inhibited from attacking brain tumor cells in animals with high levels of CaMKK2. Among mice bred without CaMKK2, T-cells remained aggressive cancer killers.
Not only did CaMMK2 impact T-cell function, but the protein also changed the behavior of macrophages, making these immune cells less helpful to T-cells. Here again, when CaMMK2 was eliminated, the macrophages aided in tumor killing.
One surprising finding was that CaMKK2, specifically in neurons, was supporting brain tumor growth and suppressing the function of the immune system. Understanding how neurons do this may identify additional therapeutic targets in the future.
Because CaMKK2 is highly expressed in tumor-supporting neurons and macrophages, the researchers said, it promotes resistance to immune-checkpoint inhibitor drugs, which turn off a cellular brake to accelerate the cancer-killing ability of T-cells.
"When mice without CaMMK2 were treated with checkpoint inhibitors, they responded and survived even longer, but mice with CaMKK2 did not," Tomaszewski said. "This suggests that CaMKK2 represents a potential therapeutic target. If we could inhibit CaMMK2, we might be able to unleash the power of the whole class of immunotherapy drugs that have been beneficial in other cancers, but not glioblastoma."
More information: William H. Tomaszewski et al, Neuronal CaMKK2 promotes immunosuppression and checkpoint blockade resistance in glioblastoma, Nature Communications (2022). DOI: 10.1038/s41467-022-34175-y
Journal information: Nature Communications
Provided by Duke University