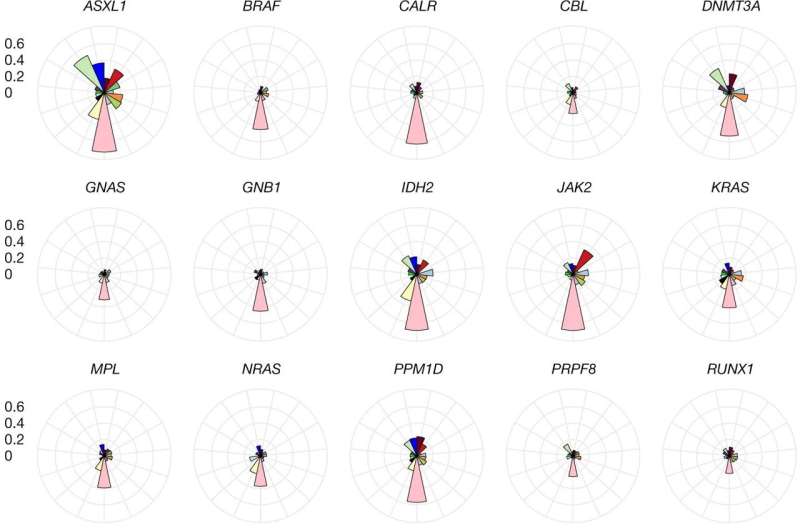
Phenome association profiles per CHIP subtype. Profiles are shown for each CHIP gene subtype reflecting phenome-wide association results. The y-axis (concentric circles) represents the proportion of phenotypes within a trait category that were nominally associated (P ≤ 0.05) with carrier status of the CHIP gene. A CHIP gene had to have at least one disease category with the proportion of associated phenotypes ≥ 0.2 to be included in the figure. As expected, haematological traits show the largest proportion of phenotypic trait associations overall. The largest number of cancer associations are seen for DNMT3A CHIP, whereas JAK2 CHIP shows the highest proportion of cardiovascular associations. Respiratory associations are most pronounced for ASXL1 CHIP. SUZ12 CHIP shows a unique profile across CHIP subtypes, with a higher proportion of ophthalmological and endocrine associations. Association models were run with age, age2, sex and age × sex, and ten ancestry-informative principal components as covariates. Credit: Nature (2022). DOI: 10.1038/s41586-022-05448-9
A team of researchers at Regeneron Pharmaceuticals has identified new genomic variants associated with clonal hematopoiesis of indeterminate potential (CHIP).
In their paper published in the journal Nature, the group describes how they used exome-wide and genome-wide association analyses to study differences in the blood of some people with somatic mutations. Nature has also published a Research Highlights piece in the same journal issue, discussing the work done by the New York team.
Hematopoiesis is a process that results in the formation of cellular blood components. And clonal hematopoiesis is the part of the process that is involved in the development of cell lineages. The importance of the overall process is highlighted by the fact that every person produces approximately 300 billion new blood cells every single day of their life.
Prior research has suggested that there are variants associated with clonal hematopoiesis of indeterminate potential in certain people—each of which can have a unique impact. In this new effort, the team at Regeneron sought to find some of them by studying information held in very large datasets, such as the UK Biobank and the Geisinger MyCode Community Health Initiative.
To find the variants they were after, the researchers focused their search efforts on 23 genes that have already been associated with CHIP. By searching through data on 628,388 individuals, they were able to identify 40,208 carriers of at least one variant associated with CHIP. They then conducted exome-wide and genome-wide studies of the carriers they had identified. In so doing, they were able to identify 24 loci—21 of which had not been seen before.
The researchers also found that they were able to identify some variants that could be associated with clonal hematopoiesis and the length of telomeres in certain individuals.
In another part of their study, the team analyzed health traits of people listed in the UK Biobank looking for associations between people who had CHIP variants and other issues. In so doing, they found associations between people who had clonal hematopoiesis variants and diseases such as COVID-19, heart problems, obesity and problems clearing infections of various types. They also found associations between individuals with CHIP and development of cancerous tumors and myeloid leukemias.
More information: Michael D. Kessler et al, Common and rare variant associations with clonal haematopoiesis phenotypes, Nature (2022). DOI: 10.1038/s41586-022-05448-9
Kirsty Minton, CHIPping away at the genetic aetiology of clonal haematopoiesis, Nature Reviews Genetics (2022). DOI: 10.1038/s41576-022-00565-7
Journal information: Nature , Nature Reviews Genetics
© 2022 Science X Network