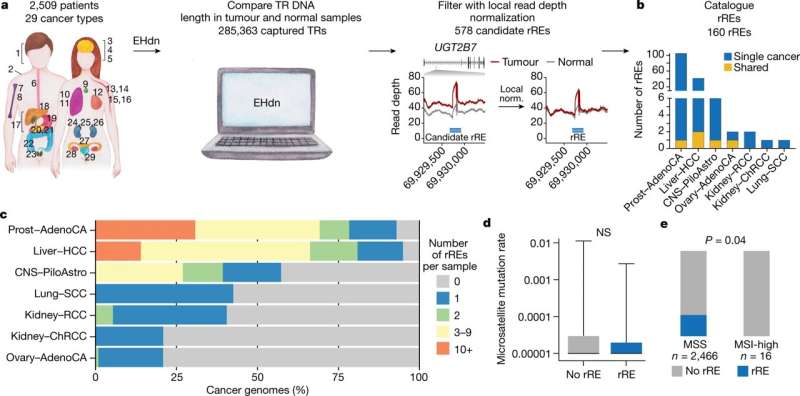
Genome-wide detection of rREs in cancer genomes. a, Scheme of the method to identify rREs in 2,509 patients across 29 human cancer types: 1, head and neck squamous cell carcinoma (Head−SCC); 2, skin–melanoma; 3, glioblastoma (CNS–GBM); 4, medulloblastoma (CNS−Medullo); 5, pilocytic astrocytoma (CNS–PiloAstro); 6, oesophageal adenocarcinoma (Oeso−AdenoCA); 7, osteosarcoma (Bone−Osteosarc); 8, leiomyosarcoma (Bone−Leiomyo); 9, thyroid adenocarcinoma (Thy–AdenoCA); 10, lung adenocarcinoma (Lung−AdenoCA); 11, lung squamous cell carcinoma (Lung−SCC); 12, mammary gland adenocarcinoma (Breast−AdenoCA); 13, B cell non-Hodgkin lymphoma (Lymph−BNHL); 14, chronic lymphocytic leukemia (Lymph−CLL); 15, acute myeloid leukemia (Myeloid−AML); 16, myeloproliferative neoplasm (Myeloid−MPN); 17, biliary adenocarcinoma (Biliary–AdenoCA); 18, hepatocellular carcinoma (Liver−HCC); 19, stomach adenocarcinoma (Stomach−AdenoCA); 20, pancreatic adenocarcinoma (Panc−AdenoCA); 21, pancreatic neuroendocrine tumor (Panc−Endocrine); 22, colorectal adenocarcinoma (ColoRect–AdenoCA); 23, prostatic adenocarcinoma (Prost−AdenoCA); 24, chromophobe renal cell carcinoma (Kidney–ChRCC); 25, renal cell carcinoma (Kidney–RCC); 26, papillary renal cell carcinoma (Kidney−pRCC); 27, uterine adenocarcinoma (Uterus−AdenoCA); 28, ovarian adenocarcinoma (Ovary−AdenoCA); 29, transitional cell carcinoma of the bladder (Bladder−TCC). b, Distribution of rREs across cancer types. c, Proportion of cancer genomes with rREs. d, STR mutation rate for cancer genomes with and without an rRE. Two-tailed Mann–Whitney test (n = 2,465 cancer genomes); NS, not significant. Boxes extend from the 25th percentile to the 75th percentile, the center line represents the median and whiskers represent minima and maxima. e, Distribution of rREs across MSS and MSI-high cancers. Chi-squared (two-tailed) test with Yates’ correction (n = 2,482 cancer genomes). Credit: Nature (2022). DOI: 10.1038/s41586-022-05515-1
A team of researchers with member affiliations across the U.S. and Canada has found evidence of multiple recurrent, repeat expansions in human cancer genomes. In their paper published in the journal Nature, the group describes searching for repeat expansions in human tumors in databanks maintained by the International Cancer Genome Consortium and The Cancer Genome Atlas group.
As the researchers note, tandem repeat (TR) in DNA sequences is known to be involved in more than 50 inheritable diseases. And as they also note, most research looking into the impact of a given TR is restricted to neurodegenerative or neurological diseases not involving cancer.
Still, some prior research has shown that some mutations are known to occur in short-tract TR sequences, which has been attributed to microsatellite instability. In this new effort, the researchers took a closer look at associations between TR sequences in cancerous tumors.
The work began by searching the International Cancer Genome Consortium and The Cancer Genome Atlas datasets, looking for somatic repeat expansions in tumor DNA for over two-dozen types of cancers. The researchers found 160 recurrent repeat expansions (rREs). They noted that all but five of those found were likely cancer subtype specifics that often appear in multiple types of common cancers, such as ovarian, kidney and lung cancers.
The researchers also conducted whole-genome long and short sequencing on two types of cancer cell lines that they had found in their original search—they studied both using Expansion Hunter Denovo (EHdn) to measure TR length. This showed that approximately 72% of the repeat expansions in the tumor tissue were in long read sequences.
The researchers also noted that prior research with a molecule known as Syn-TEF1 has been found to reduce TR in certain neurological diseases, prompting them to test its use in several cancer types. This led to reductions in cell proliferation. They suggest this finding indicates that other molecules or therapies originally intended to treat neurological diseases with TR associations may help treat some cancers as well.
More information: Graham S. Erwin et al, Recurrent repeat expansions in human cancer genomes, Nature (2022). DOI: 10.1038/s41586-022-05515-1
Journal information: Nature
© 2022 Science X Network