Rethinking how cancer cells evade targeted therapy
Glioblastomas (GBMs) are incurable brain tumors with a prognosis of about one-and-a half years on average. They are highly resistant to treatment and have defied all attempts at precision therapy.
In their study publishing December 20 in Nature Cancer, first author Lin Wang, Ph.D. and senior author Aaron Diaz, Ph.D., found that phenotype switching, as opposed to genetic evolution, may be the escape mechanism that explains the failure of precision therapies to date. They found that some cells shift to a mesenchymal, radiation-resistant phenotype (state) as a stress response following standard therapy.
"We asked if there is another mechanism that explains therapeutic resistance," said Diaz, associate professor of neurological surgery at the UCSF Weill Institute for Neurosciences. "Our study concludes that, rather than a genetic evolution, there is a phenotypic plasticity or transition which allows these cells to evade therapy."
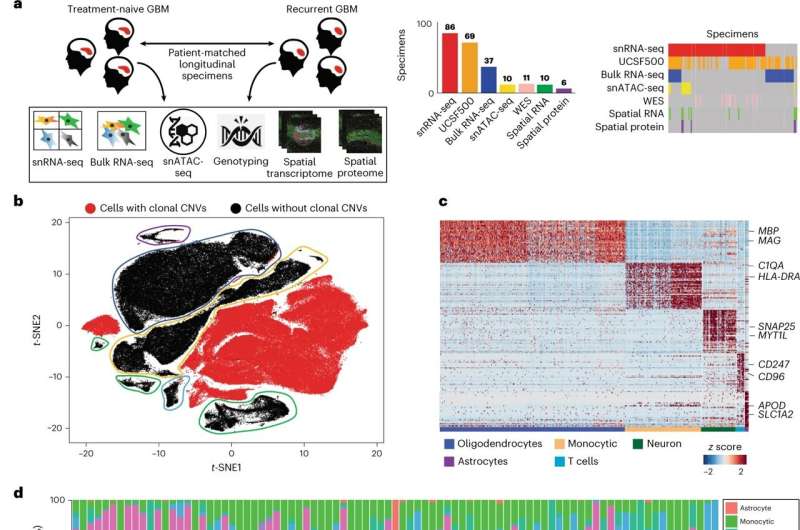
To identify what drives treatment resistance to standard therapy, as well as the cellular source of recurrent disease, UCSF researchers used single-nucleus RNA, open-chromatin, spatial profiling to analyze 86 primary-recurrent, patient-matched, paired GBM specimens. This unprecedented cohort represented decades of biobanking at UCSF.
With access to thirty years' worth of GBM's, the scientists were able to present novel cell-intrinsic and cell-extrinsic targets as well as a single-cell multi-omics atlas of GBM under therapy. This was the first time that researchers were able to comprehensively map intra-cellular signaling in the tumor-anatomical niches of recurrent GBM and identify novel cell-extrinsic therapeutic targets.
The 86 specimens contained the cellular tumor and adjacent non-malignant tissue from the surgical margin. This unique cohort enabled Diaz and his team to analyze communications between malignant and non-malignant glia. They found that cells in this surgical margin acted as niches of recurrence where non-malignant glia were broadcasting pro-growth signals that influenced the tumor cells to regrow. These paracrine (cell extrinsic) signals stimulated the activator protein (AP1) pathway, leading to mesenchymal transition, therapy resistance and tumor recurrence.
These mesenchymal transitions were apparent in the tumor samples after therapy, along with increased numbers of cycling mesenchymal cells. Pro-growth signals from the tumor microenvironment, as well as the cell-intrinsic response to radiation therapy, drove the mesenchymal shift through a stress-response pathway mediated by AP1.
The researchers were able to combine two techniques to achieve a map of paracrine signals—single-nucleus RNA-sequencing of frozen tissue specimens and spatial transcriptomics profiling of formalin-fixed specimens. From each tumor biopsy, they performed single-nucleus RNA-seq which measures transcriptome-wide gene expression in individual cells, for thousands of cells at a time.
They compared the expression of signal receptors to that of their known ligands, between the different cell populations sequenced. They then validated that cells from these populations physically interact and signal in situ, using spatial transcriptomics.
"This is the first single-cell longitudinal study of this scale in glioma," said Diaz. "It's also a study that could be done only at UCSF, because it represents decades of careful biobanking of surgical specimens. Since all the specimens came from UCSF, we know that the treatment histories are homogeneous, in that each patient received only standard-of-care therapy. It's both this cohort's scale and treatment uniformity that enable us to see past patient specific effects to the underlying biology of the disease."
More information: Lin Wang et al, A single-cell atlas of glioblastoma evolution under therapy reveals cell-intrinsic and cell-extrinsic therapeutic targets, Nature Cancer (2022). DOI: 10.1038/s43018-022-00475-x