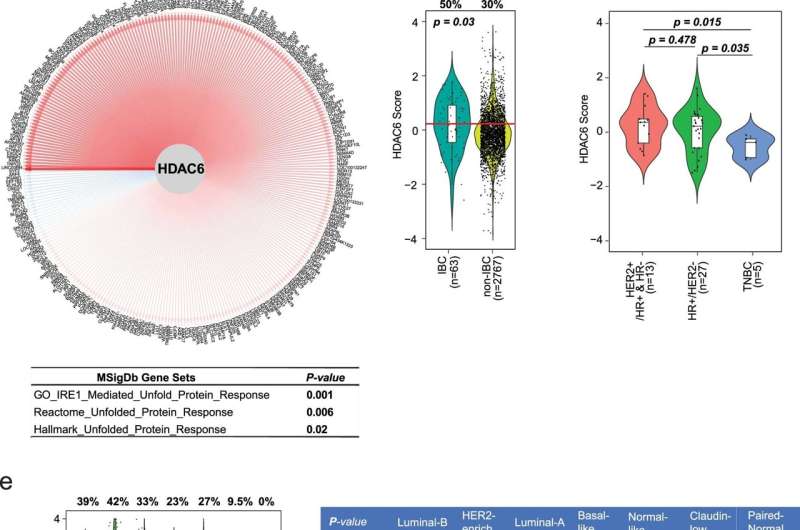
HDAC6 score in BC. (a) The number of samples and dataset of origin used to evaluate the HDAC6 regulon. (b) Overlap of the original and updated HDAC6 regulons. P value was estimated using two-tailed Fisher’s exact test. N was determined by the total number of genes for network inference. (c) The network plot of HDAC6 and updated HDAC6 regulon. Edge width is corresponding to the correlation strength measured by mutual information. Red and blue edges indicate positive and negative correlations between HDAC6 and each of the regulon genes. The table below shows the pathway enrichment of the genes in the regulon, showing its association with unfolded protein response. P value was estimated using two-tailed Fisher’s exact test. (d) New HDAC6 score comparing IBCs vs non-IBCs. (e) HDAC6 scores of all BCs from TCGA and METABRIC are divided into molecular subtypes. (f) HDAC6 scores in 45 ductal metastatic breast cancer samples from the MBC Project divided into histological molecular subtypes. In d, e and f, the center line indicates the median value. The lower and upper hinges represent the 25th and 75th percentiles, respectively, and whiskers denote 1.5x interquartile range; the red line represents the median of the HDAC6 scores in IBC samples and the numbers over each whisker plot indicate the percentage of samples over this value in each clinical subtype. Sample size (n = number of samples) of each group was indicated in the axis labels. P value was estimated using two-tailed t test. Credit: Nature Cancer (2022). DOI: 10.1038/s43018-022-00489-5
A team of researchers affiliated with a host of medical institutions across the U.S. has found that the HDAC6 score can be used to predict the response of some types of cancer to administration of the histone ricolinostat.
In their paper published in the journal Nature Cancer, the group studied information on breast cancer patients who had received ricolinostat. Joschka Hey, Maria Llamazares Prada and Christoph Plass, with the German Cancer Research Center, published a News & Views piece in the same journal issue outlining current research into identifying cancer patients who may benefit from certain treatments and the work by the team in this new effort.
The HDAC6 score is a number derived through measuring expression of histone deacetylase 6, an enzyme that is encoded by the HDAC6 gene in humans. It has been developed as a score in association with treating cancer. In this new effort, the researchers used the score as a means of measuring response to a drug meant to treat HR+/HER2- breast cancer patients—the histone deacetylase 6 inhibitor ricolinostat.
In their work, the researchers focused their efforts on patients with inflammatory breast cancer, a rare subtype that accounts for a small number of cases. To determine the impact of ricolinostat on such patients, the researchers applied the HDAC6 score to tumor data in four major datasets containing large amounts of cancer patient data.
They found that approximately 30% of all the types of breast cancers they tested suggested a response to HDAC6 inhibition, which further suggests that the HDAC6 score may be used as a marker for a positive response to administration of ricolinostat in breast cancer patients.
The researchers found a response in 14 breast cancer lines and also in some mouse models and have therefore begun drawing up plans for conducting a Phase 1b study. They also note that administration of the drug nab-paclitaxel would be included in such a study because it was the drug predominantly given with ricolinostat in the patients documented in the datasets.
More information: Tizita Z. Zeleke et al, Network-based assessment of HDAC6 activity predicts preclinical and clinical responses to the HDAC6 inhibitor ricolinostat in breast cancer, Nature Cancer (2022). DOI: 10.1038/s43018-022-00489-5
Joschka Hey et al, HDAC6 score: to treat or not to treat?, Nature Cancer (2022). DOI: 10.1038/s43018-022-00494-8
Journal information: Nature Cancer
© 2023 Science X Network