This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Heart attack study could change the game in regenerative medicine
Sanford Burnham Prebys researchers have identified a group of proteins that could be the secret to cellular reprogramming, an emerging approach in regenerative medicine in which scientists transform cells to repair damaged or injured body tissues. The researchers were able to reprogram damaged heart cells to repair heart injuries in mice following a heart attack. The findings, which appear in the journal Nature Communications, could one day transform the way we treat a variety of diseases, including cardiovascular disease, Parkinson's and neuromuscular diseases.
"Even if a person survives a heart attack, there could still be long-term damage to the heart that increases the risk of heart problems down the line," says lead author Alexandre Colas, Ph.D., an assistant professor in the Development, Aging and Regeneration Program at Sanford Burnham Prebys. "Helping the heart heal after injury is an important medical need in its own right, but these findings also pave the way for wider applications of cell reprogramming in medicine."
Even though each of our cells has the same number of genes—approximately 20,000—cells can select which genes to "turn on" and "turn off" to change what they look like and what they do. This is the foundation of cellular reprogramming.
"Cellular reprogramming could, in theory, allow us to control the activity and appearance of any cell," says Colas. "This concept has huge implications in terms of helping the body regenerate itself, but barriers to reprogramming mechanisms have prevented the science from moving from the lab to the clinic."
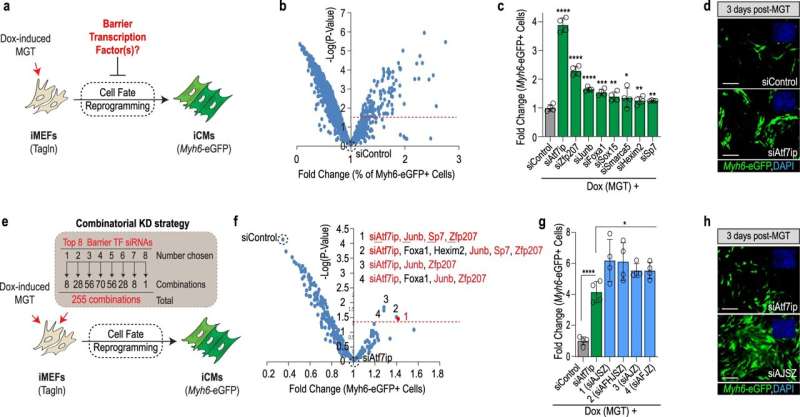
The researchers identified a group of four proteins, named AJSZ, that help solve this problem. "By blocking the activity of these proteins, we were able to reduce scarring on the heart and induce a 50% improvement in overall heart function in mice that have undergone a heart attack," says Colas.
Although the researchers were primarily focused on heart cells, they determined that AJSZ is universal to all cell types. This suggests that targeting AJSZ could be a promising treatment approach for a variety of human diseases.
"This is helping us solve a very big problem that a lot of researchers are interested in," says Colas. "Even more important, this breakthrough is a significant step forward on our way to turning these promising biological concepts into real treatments."
The next steps in translating their discovery into a potential treatment is to explore different ways of blocking the function of the AJSZ proteins. According to Colas, the most promising option would be to use a small molecule drug to block the activity of AJSZ.
"We need to find a way to inhibit these proteins in a way we can control to make sure we are only reprogramming the cells that need it," says Colas. "We will be screening for drugs that can help us inhibit these proteins in a controlled and selective manner in the coming months."
More information: Maria A. Missinato et al, Conserved transcription factors promote cell fate stability and restrict reprogramming potential in differentiated cells, Nature Communications (2023). DOI: 10.1038/s41467-023-37256-8