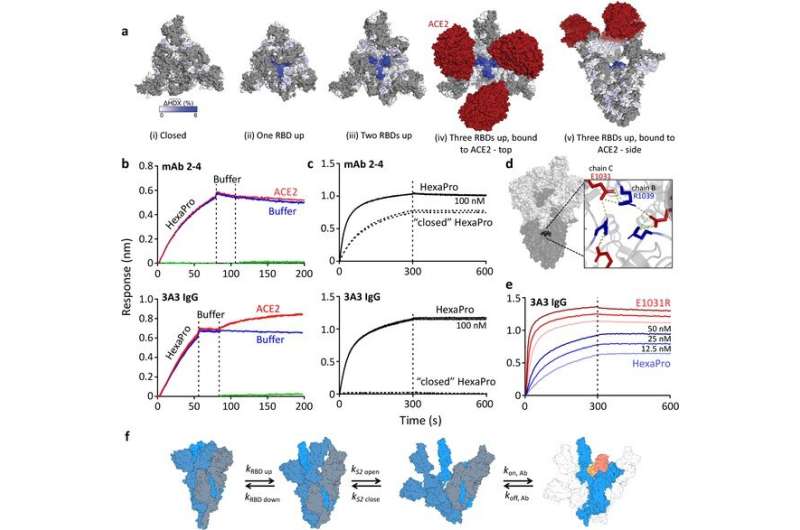
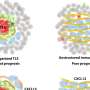
The hinge epitope is accessible only in an RBD-up and S2-open spike conformation. (a) Trimeric SARS-2 spike in various conformations colored according to difference in deuterium fractional uptake between SARS-2 HexaPro spike alone and with 3A3 IgG. The hinge epitope within S2 is colored dark blue in structures of wild-type SARS-2 spike in the (i) three RBDs down or closed conformation (PDB: 6XR8) and in structures of stabilized spike with (ii) one RBD up (PDB: 6VSB), (iii) two RBDs up (PDB: 7A93), or (iv) three RBDs up while bound to ACE2 (red) in top-view and (v) sideview (PDB: 7A98). Residues lacking coverage in the HDX experiment are indicated in gray. (b) Antibody 3A3 (bottom) or control mAb 2–4 (top) were coupled to anti-Fab BLI sensors and allowed to capture HexaPro or nothing (buffer, green line), then dipped into buffer (baseline), and finally dipped into ACE2-Fc (ACE2, red) or nothing (buffer, blue). (c) BLI binding of immobilized control mAb 2–4 (top) or antibody 3A3 (bottom) to 100 nM HexaPro (solid) or HexaPro locked into the ‘closed’ conformation (dashed). Vertical dashed lines indicate start of dissociation phase. (d) The network of hydrogen bonds formed by residues E1031 and R1039 across protomers deep in the S2 core is shown on intact HexaPro spike and in detail in a top view (PDB: 6XKL). (e) Antibody 3A3 was coupled to anti-Fc BLI sensors and allowed to bind HexaPro or E1031R HexaPro (E1031R) spike protein. All BLI data are representative of biological duplicates. Each experiment was repeated in technical duplicate except e, which was tested once at each concentration to allow all data to be collected simultaneously for direct comparison. (f) Model of the kinetic changes required for antibody binding to the hinge epitope, including conversion of the RBDs into the up position and some degree of opening of the S2 domain in addition to typical antibody association and dissociation kinetics (generated using PDB 6XV8 and 7A98). Credit: eLife (2023). DOI: 10.7554/eLife.83710
Researchers searching for new therapeutic targets in the SARS-CoV-2 virus have identified a potential Achilles' heel that exists in all coronaviruses, according to a study published today in eLife.
The research could aid the development of more powerful antibody drugs and vaccines against the virus that currently causes COVID-19 and might also protect against emerging coronaviruses that could cause future pandemics.
Most vaccines and antibody-based treatments for COVID-19 neutralize the SARS-CoV-2 virus by disrupting interactions between the protein spike on the virus and the ACE2 receptor on human cells, which the virus hijacks to gain entry. But mutations in the spike protein mean that emerging variants of SARS-CoV-2 can escape the human antibody response, so that treatments do not work and vaccinated individuals experience breakthrough infections. One way to address this issue is to target treatments and vaccines against parts of the spike protein that the virus needs for survival and cannot mutate.
The SARS-CoV-2 spike protein is made up of two subunits—called S1 and S2. The S1 subunit contains a region that binds to the ACE2 receptor, while the S2 subunit allows the virus to fuse with the membrane of the cell it is gaining access to. Most mutations in the spike protein affect its S1 domain, but the S2 domain is highly conserved across all seven human coronaviruses, suggesting that it could be a good target for therapeutic antibodies and vaccines.
"We know that S2-directed antibodies are produced following a COVID-19 infection, and similar subunits in flu and HIV are also targeted by the body's antibodies," explains Rui Silva, a former graduate student at The University of Texas at Austin, U.S., who led the work in close collaboration with fellow graduate student Yimin Huang and Senior Scientist Annalee Nguyen.
"However, we know very little about the antibodies that bind the S2 subunit. Fewer than five percent of the roughly 7,000 known anti-SARS-CoV-2 antibodies bind S2 and only two classes of S2 binding antibodies have been characterized in detail. We have identified a third class of S2 antibody that binds a highly conserved part of the S2 subunit."
The team isolated the antibody by immunizing mice with the S2 subunit from the related Middle Eastern respiratory syndrome virus and collected the antibodies produced that bound spike from this virus as well as SARS-CoV-2. They then analyzed the most promising anti-S2 antibody further.
They found that this antibody binds a hinge in the S2 subunit that plays a crucial role when the subunit changes its shape during the virus's fusion with the human cell membrane. Further analysis showed that access to the point where the antibody binds to this hinge (called the epitope) depended on the shape-shifting dynamics of the overall spike protein; when the spike is bound to the ACE2 receptor, the epitope on the hinge is accessible.
The team then conducted a series of experiments to see if targeting this hinge epitope had any impact on viral activity. They found that although S2-directed antibodies could prevent the spike protein's ability to fuse viral and human cell membranes, they were less potent at neutralizing the virus altogether.
However, as well as directly neutralizing viruses, antibodies also indirectly kill virus-infected cells, by triggering other processes. In one experiment, S2-directed antibodies could trigger human natural killer cells to destroy SARS-CoV-2 infected cells. In another, the S2 antibodies caused cells called monocytes to gobble up and destroy the infected cells. This suggests that although S2 antibodies might not be strong enough to directly block infection, they could be used to help boost or protect an immune response.
"We have identified an epitope in the S2 subunit of SARS-CoV-2 that is highly conserved across all pathogenic coronavirus strains," concludes senior author Jennifer Maynard, ZD Bonner Professor of Chemical Engineering at The University of Texas at Austin. "Although targeting this epitope alone is unlikely to be potent enough as a treatment or vaccine, therapeutic strategies that can enhance access to this epitope could allow existing human antibodies to more effectively promote viral clearance through other antibody-directed cell-killing processes."
More information: Rui P Silva et al, Identification of a conserved S2 epitope present on spike proteins from all highly pathogenic coronaviruses, eLife (2023). DOI: 10.7554/eLife.83710
Journal information: eLife
Provided by eLife