This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Genetic clues could predict leukemia patients' risk of treatment failure
New research led by UCL and Great Ormond Street Hospital (GOSH) to predict which childhood leukemia patients are at higher risk of not responding well to chemotherapy could allow clinicians to refine treatment strategies to give the best chance of success.
The study, published in the Journal of Clinical Oncology, combined U.K. trial data from 2003–2019 to see which patients had worse outcomes. They then used whole genome sequencing to look for genetic clues that could be used to predict this risk in future patients.
The team hope that these genetic clues can be used to identify high-risk patients early, who may be eligible to take part in trials for the latest immunotherapies, such as CAR-T cell therapy.
Acute lymphoblastic leukemia (ALL) is the main type of childhood leukemia and is split into two subtypes, B-Cell ALL (B-ALL) and T-Cell ALL (T-ALL). The current conventional therapies for ALL include induction chemotherapy and bone marrow transplant. But patients are three times more likely to not respond well to therapy and suffer poor outcomes in T-ALL compared with B-ALL, and there are fewer treatment options available when patients relapse.
This may be about to change, with trials underway for highly promising immunotherapies such as CAR-T cell therapy. This technique was recently used by scientists at UCL and GOSH to treat a patient's "incurable" T-ALL leukemia after conventional treatments had failed. Though the trial continues, the patient is still cancer free almost a year after starting treatment.
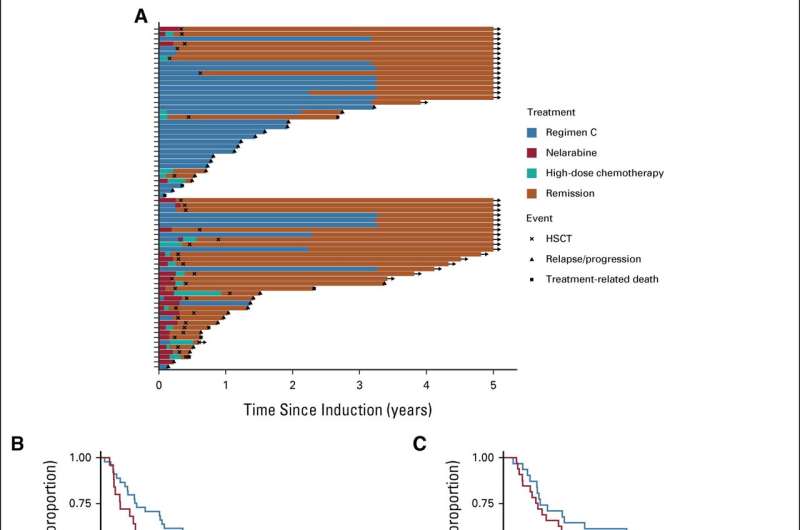
In the new study, the team analyzed data from two clinical trials that involved ALL patients treated in the U.K. between 2003 and 2019, to see what happened to the patients and identify the risk factors associated with conventional treatment failure.
Frozen cells from these trial patients were then whole genome sequenced in order to look for genetic markers associated with poor outcomes, and which might be used to predict the risk of conventional treatment failure in future patients.
The researchers found that one in five older patients, in this case teenagers and young adults, experienced poor outcomes compared to just one in ten younger patients.
Though the genetic profile of the cancers they studied was complex, with individual cancers often very different from one another, the researchers found that mutations in the TAL1, MYC and RAS molecular pathways could be used to quantify a patient's risk of treatment failure.
Dr. David O'Connor (UCL Cancer Institute and Great Ormond Street Hospital) said, "Genetic stratification, where you analyze a patient's genome to assess their risk of certain outcomes, has been successfully used to improve B-ALL treatment. But until now we haven't had the data to do this for T-ALL. Our results could identify a group of high-risk patients who are less likely to respond well to chemotherapy. All childhood leukemia patients in the U.K. undergo whole genome sequencing when they are diagnosed, so the genetic information should already be available to do this."
Genetic stratification of patients to identify those who are less likely to respond well to conventional treatments would mean that they could be considered for alternatives, such as the CAR-T cell therapies that are now available in clinical trials.
Professor Marc Mansour (UCL Cancer Institute and UCL Great Ormond Street Institute of Child Health) said, "Around 10% of pediatric T-ALL patients do not respond to initial chemotherapy and the chance of relapse is twice as high as in B-ALL. This is compounded by the fact that there are fewer treatment options for relapsed T-ALL patients. So in T-ALL it's even more important to provide the treatment that will give the highest chances of remission. The genetic markers that we've identified in this study will allow us to provide the best treatment strategy."
More information: David O'Connor et al, The Clinicogenomic Landscape of Induction Failure in Childhood and Young Adult T-Cell Acute Lymphoblastic Leukemia, Journal of Clinical Oncology (2023). DOI: 10.1200/JCO.22.02734