Chemists at the University of California, San Diego have produced the first high resolution structure of a molecule that when attached to the genetic material of the hepatitis C virus prevents it from reproducing.
Hepatitis C is a chronic infectious disease that affects some 170 million people worldwide and causes chronic liver disease and liver cancer. According to the Centers for Disease Control and Prevention, hepatitis C now kills more Americans each year than HIV.
The structure of the molecule, which was published in a paper in this week's early online edition of the journal Proceedings of the National Academy of Sciences, provides a detailed blueprint for the design of drugs that can inhibit the replication of the hepatitis C virus, which proliferates by hijacking the cellular machinery in humans to manufacture duplicate viral particles.
Finding a way to stop that process could effectively treat viral infections of hepatitis C, for which no vaccine is currently available. But until now scientists have identified few inhibiting compounds that directly act on the virus's ribonucleic acid (RNA) genome—the organism's full complement of genetic material.
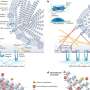
"This lack of detailed information on how inhibitors lock onto the viral genome target has hampered the development of better drugs," said Thomas Hermann, an associate professor of chemistry and biochemistry at UC San Diego who headed the research team, which also included scientists from San Diego State University. The team detailed the structure of a molecule that induces the viral RNA to open up a portion of its hinge-like structure and encapsulate the inhibitor like a perfectly fit glove, blocking the ability of the hepatitis C virus to replicate.
The molecule is from a class of compounds called benzimidazoles, known to stop the production of viral proteins in infected human cells. Its three-dimensional atomic structure was determined by X-ray crystallography, a method of mapping the arrangement of atoms within a crystal, in which a beam of X-rays strikes a crystal and causes the beam of light to spread. The angles and intensities of the light beams allowed the scientists to calculate the structure of the viral RNA-inhibitor complex.
"This structure will guide approaches to rationally design better drug candidates and improve the known benzimidazole inhibitors," said Hermann. "Also, the crystal structure demonstrates that the binding pocket for the inhibitors in the hepatitis C virus RNA resembles drug-binding pockets in proteins. This is important to help overcome the notion that RNA targets are so unlike traditional protein targets that drug discovery approaches with small molecule inhibitors are difficult to achieve for RNA."
Provided by University of California - San Diego