(Medical Xpress)—A large team of researchers with members from several universities and hospitals in the U.S. has developed a sequencing platform that generates "clinical-grade" data using whole exome sequencing (WES) from small amounts of DNA extracted from clinical tumor samples. In their paper published in the journal Nature Medicine, the team describes their new method and how it might soon be used in routine clinical practice.
Over the past several years scientists have learned that different types of cancers with different types of associated tumors can be genetically different from one another and that the different types respond differently to different treatment options. Because matching the type of treatment options to tumor types is still so new, researchers are still working out the best ways to go about doing so. Quite often, small samples of tumors are taken which are then genetically tested to see if the genome, or parts of it match known conditions that have been successfully treated using a certain therapy. More recently, it's become clear that WES offers a better approach, it allows for sequencing the whole genome rather than just a part of it. Efforts to do so have, however, been held back by the lack of a method to perform such sequencing on tumor samples stored using fixed in formalin and embedded in paraffin (FFEP), the standard method used to store such samples—it tends to degrade genetic material. In this new effort the researchers came up with a way to use such samples to perform WES.
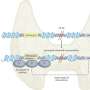
The breakthrough was the development of an algorithm that is run on a computer that analyses the results of WES—they've named it precision heuristics for interpreting the alteration landscape (PHIAL). It collects genetic data and then sorts it by variation based on already known information about such variations and treatment options for them. It finishes by noting what has been found regarding the genome and by noting treatment options that have been the most successful at treating tumors with similar variations.
The researchers tested their system on 511 actual patients and found it was able to pick out 1,842 alterations out of 250,000 variants for which there are known treatment options. They also found that the system was able to identify clinically relevant alterations in 15 of 16 patients, one of whom was a lung cancer patient who responded positively to a particular therapy after being guided there by the system.
The researchers note that their system allows for expanding the WES knowledgebase—data from their system can be added to an ever increasing database, providing the algorithm ever more information to use in sorting.
More information: (Medical Xpress)—Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine, Nature Medicine (2014) DOI: 10.1038/nm.3559
Abstract
Translating whole-exome sequencing (WES) for prospective clinical use may have an impact on the care of patients with cancer; however, multiple innovations are necessary for clinical implementation. These include rapid and robust WES of DNA derived from formalin-fixed, paraffin-embedded tumor tissue, analytical output similar to data from frozen samples and clinical interpretation of WES data for prospective use. Here, we describe a prospective clinical WES platform for archival formalin-fixed, paraffin-embedded tumor samples. The platform employs computational methods for effective clinical analysis and interpretation of WES data. When applied retrospectively to 511 exomes, the interpretative framework revealed a 'long tail' of somatic alterations in clinically important genes. Prospective application of this approach identified clinically relevant alterations in 15 out of 16 patients. In one patient, previously undetected findings guided clinical trial enrollment, leading to an objective clinical response. Overall, this methodology may inform the widespread implementation of precision cancer medicine.
Journal information: Nature Medicine
© 2014 Phys.org