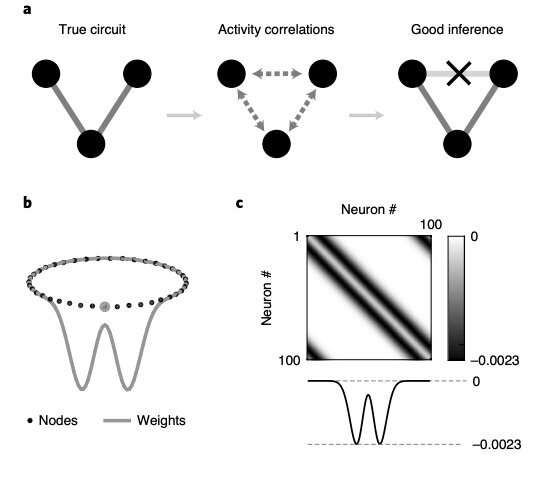
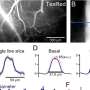
A) Left: figure representing a three-neuron circuit with two connections. Center: all neurons are correlated. Right: circuit inference algorithms should ‘explain away’ correlations between the top neurons based on their common input, ruling out a direct connection. B) Neurons (black dots) in a ring network. Gray curve, Mexican hat-shaped weights from an example node (gray dot) to the rest; all other neurons have the same weight profile. C) The resulting weight matrix W (top) is circulant, comprising rotations of the same row (bottom). D) Scalar parameter r modulates the strength of all recurrent weights. Spike raster plots (top) and snapshots of synaptic activity (bottom) of the network at weak (red) and strong (blue) weights (small and large r). Credit: Das & Fiete, Nature Neuroscience (2020).
In recent years, a growing number of computer scientists have tried to develop computational methods inspired by the structure, function and plasticity of neural circuits in the human brain. Achieving a comprehensive understanding of biological neural circuits is of vital importance for the creation of these neuro-inspired computing systems.
To fully comprehend the mechanisms that allow biological neural circuits to compute information and adapt over time, neuroscientists should be able to examine the connections between individual neurons. While recent advancements in circuit tracing techniques have opened up new possibilities for studying these connections, collecting data using these techniques can still be very challenging and expensive.
Some scientists have thus devised statistical methods of estimating neural connectivity based on multicell neural activity recordings. While these methods are widely used, they might not lead to reliable representations of neural connections.
Researchers at the University of Texas at Austin have recently carried out a study investigating the effectiveness of existing methods for algorithmically estimating the wiring of neural networks. Their findings, published in Nature Neuroscience, suggest that even the most sophisticated among these methods are biased and tend to infer connections between neurons that are not actually connected, but rather highly correlated.
"Because it is difficult to directly measure the wiring diagrams of neural circuits, there has long been an interest in estimating them algorithmically from multicell activity recordings," the researchers explain in their paper. "We show that even sophisticated methods, applied to unlimited data from every cell in the circuit, are biased toward inferring connections between unconnected but highly correlated neurons. This failure to 'explain why' connections occurs when there is a mismatch between the true network dynamics and the model used for inference, which is inevitable when modeling the real world."
To assess the effectiveness of statistical methods to infer neural connectivity, the researchers constructed a series of recurrent networks with varying absolute recurrent weight strengths, but with the same network architecture. The different recurrent weight strengths they used moved the neural circuit across different regimes, namely weak (i.e., sensory), medium (amplifying sensory) and strong (memory) recurrent regimes.
In the last of these regimes, when recurrent weights were strong, they observed the emergence of a large amount of neural activity patterns, with algorithmic methods finding correlations between neurons that were, in fact, unconnected. Interestingly, the researchers found that the same type of error arises when using a wide range of algorithmic methods for estimating neural connectivity based on multicell brain activity recordings. In their paper, they thus emphasize the need to be particularly careful when trying to infer casual relationships between variables based on statistical models that calculate correlations.
While the findings they collected reiterate the challenges associated with the examination and estimation of neural connectivity, the researchers also highlight the potential of a few emerging techniques, including recent advances in the field of connectomics and methods that involve the perturbation and consequent monitoring of neurons. Finally, they introduce a new approach based on far-out-of-equilibrium sampling that could be complementary to these two emerging methods, as it combines their strengths and advantages.
"Our results suggest that, with enough out-of-equilibrium data, performing even a simple correlational inference could provide much better estimates of the true connectivity in recurrent networks than using sophisticated inference algorithms on equilibrium data," the researchers write in their paper.
The study could soon inspire new research exploring the potential of techniques for estimating the structure and connectivity of neural architectures. In addition, the far-out-of-equilibrium sampling approach could potentially prove useful for studying neural mechanisms other than neural connectivity, such as neural and synaptic adaptation.
More information: Abhranil Das et al. Systematic errors in connectivity inferred from activity in strongly recurrent networks, Nature Neuroscience (2020). DOI: 10.1038/s41593-020-0699-2
Journal information: Nature Neuroscience
© 2020 Science X Network