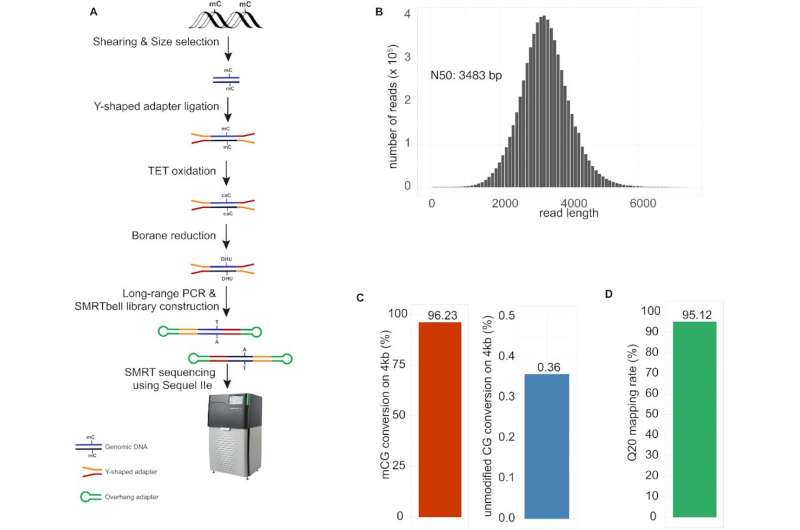
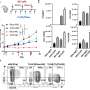
Development of wglrTAPS. (A) Schematic representation of the wglrTAPS. (B) Sequence length distribution of wglrTAPS HiFi read. (C) Conversion rate of wglrTAPS at methylated CpG sites and false-positive rate of wglrTAPS at non-methylated CpG sites from CmCGG-methylated 4-kb spike-in. (D) Fraction of mapped reads with ≥Q20 in wglrTAPS. Credit: Nucleic Acids Research (2022). DOI: 10.1093/nar/gkac612
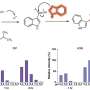
In a new paper published in Nucleic Acids Research, a team led by Ludwig Oxford's Chunxiao Song reported a method for whole-genome long-read sequencing using TAPS (for Tet-assisted pyridine borane sequencing), the method his and Benjamin Schuster-Böckler's groups published in 2019 for the detection of cytosine methylation and hydroxymethylation.
Unlike other DNA methylation sequencing methods, TAPS does not rely on the harsh chemical bisulphite, which degrades a lot of the DNA sample. This has allowed Song's group to adapt the method for applications such as liquid biopsies. Long-read sequencing, meanwhile, permits the mapping of repetitive and complex genomic regions, which can reveal new information about the genome. Song and his team show that whole-genome long-read TAPS uncovered many sites of methylation that were not present in data from short-read TAPS. The method also enabled the detection of allele-specific methylation in imprinting genes.
More information: Jinfeng Chen et al, Whole-genome long-read TAPS deciphers DNA methylation patterns at base resolution using PacBio SMRT sequencing technology, Nucleic Acids Research (2022). DOI: 10.1093/nar/gkac612
Journal information: Nucleic Acids Research
Provided by Ludwig Cancer Research