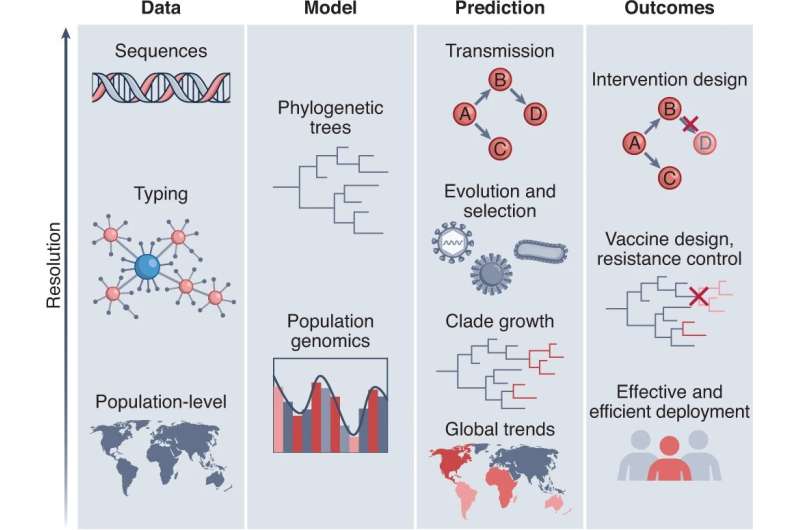
Data, models, predictions and outcomes may cover multiple levels of resolution. For example, data may comprise sequences, from which we wish to model sequence types or genotypes, to forecast global disease trends and thereby design an efficient resource deployment strategy. Alternatively, the composition of the pathogen population by larger sub-populations (for example, serotype or variant) may be the focal level for genetic diversity. There are myriad combinations, from which the scientist must determine the optimum scale at each step. Credit: Nature Microbiology (2022). DOI: 10.1038/s41564-022-01233-6
Simon Fraser University researchers are advocating for the inclusion of genomic data into forecasting models to better understand the spread of infectious diseases. The researchers say incorporating this data into forecasting models can inform monitoring, coordination and help determine where resources are needed.
In a paper published this week in Nature Microbiology, SFU mathematics researchers Caroline Colijn, Pengyu Liu and Jessica Stockdale note that genomic sequencing technologies have improved to the point where it is possible to consistently sample over time to understand how pathogens mutate and evolve to produce new variants or strains.
As the technology has improved, they say it has become more feasible to integrate genomic data from viruses and other pathogens into predictive mathematical models that forecast the spread of an infection. Models could incorporate the pathogen's diversity, the rate of transmission and interventions such as antibiotic treatment of vaccination.
"Genomic data can be used to make forecasting more accurate in assessing risks of immune evasion and antimicrobial resistance (resistance to antibiotics or other treatment options) and can help in mitigating those risks," says Colijn, who holds a Canada 150 Research Chair in Mathematics for Evolution, Infection and Public Health. She also oversees the Canadian Network for Modeling Infectious Disease (CANMOD) and is a Scientific Co-director of the Pacific Institute on Pathogens, Pandemics and Society (PIPPS) based at SFU.
Previous research has used genomic data collected over the past 20 years to analyze the evolution and spread of diseases in retrospect. The team suggests that incorporating genomic data into predictive models could allow for more accurate forecasting of pathogen behavior into the future. Genomic data would be combined with other sources of information such as epidemiological, clinical and surveillance system data.
The team adds that that there may be ethical considerations that need to be taken into account when using genomic data, and challenges to overcome with in developing the methods to integrate modeling, forecasting and genomics.
The researchers emphasize the importance of accurate modeling to project the possible future burden of disease, understand the likely patterns of evolution and diversity in pathogens and inform decisions about infectious disease control.
Gaining an understanding of how a virus behaves can also inform communication campaigns that advise the public of actions to take that will reduce their risk of infection. The World Health Organization also relies on models to estimate disease burden, make projections and inform policy.
More information: Jessica E. Stockdale et al, The potential of genomics for infectious disease forecasting, Nature Microbiology (2022). DOI: 10.1038/s41564-022-01233-6
Journal information: Nature Microbiology
Provided by Simon Fraser University