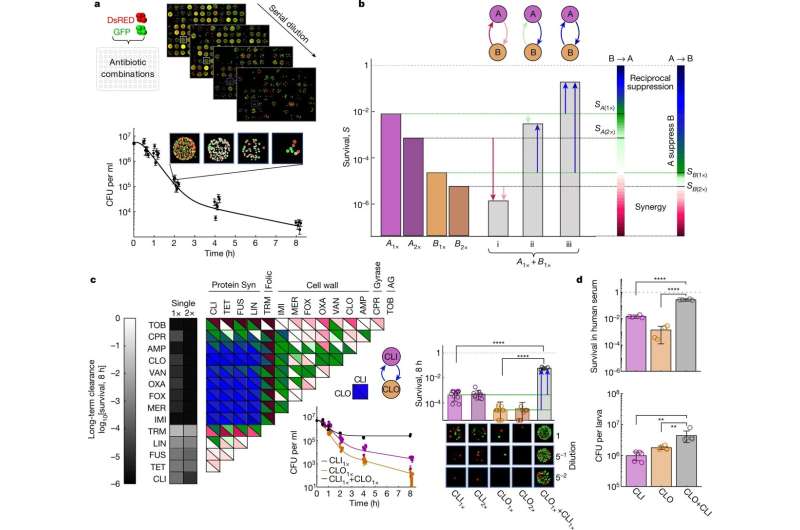
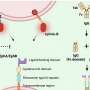
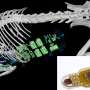
Systematic quantification of pairwise drug interactions reveals emergent reciprocal suppression in long-term population clearance. a, Cell viability over time after antibiotic treatment was measured by high-throughput microplating and automated image analysis of a mixed DsRed- and GFP-tagged S. aureus strain. Example data are shown for treatment with CLI (n = 6 wells; fivefold serial dilution microplate images are shown for one replicate at one time point). b, Schematic of directional interactions. Contrasting survival after a combined treatment with two drugs, A and B, at a fixed cidal concentration (Supplementary Table 1, gray bars, SA+B); the survival under drugs A and B alone at the same or double the concentration (SA(1×), SA(2×) (purple bars); SB(1×), SB(2×) (orange bars)) defines two-directional interaction scores for the effect of drug B on drug A (B→A, left color scale) and the effect of A on B (A→B, right color scale). The combined effect could be synergistic (SA+B < min[SA(2×), SB(2×)]) (i); non-reciprocal suppressive (SB(1×) < SA+B < SA(1×), A suppresses B) (ii); or reciprocal suppressive (SA+B > max[SA(1×), SB(1×)]) (iii). c, Measurements of directional interaction scores for pairwise combinations of 14 antibiotics in long-term clearance efficacy (2 strains were measured in A1×, A2× and A1× + B1× in 6, 4 and 2 experiments, respectively). The top (bottom) triangles show the interaction score for the effect of the drugs in the row (column) on the drugs in the column (row). Inset: the killing curves and final survival fraction for a representative suppressive drug combination (CLI + CLO). Statistical analysis was performed using z-tests; P < 10−10. Points on the axis are below the detection limit. For each treatment, representative microplating images of one replicate are shown. d, Bacteria viability after 8 h exposure to CLI–CLO in human serum (top; 2 strains were measured in 2 experiments; P < 10−10, two-tailed z-tests) and in G. mellonella larvae (d, bottom; n = 10 AROLarvae, homogenized and plated in 5 pairs; from left to right, P = 0.008 and P = 0.008, two-tailed Mann–Whitney U-tests). For a, c and d, the error bars represent the 95% confidence intervals calculated from the colony counts by the Poisson’s model (a and c (middle)) or 2× s.e.m. (c (right) and d). Credit: Nature (2022). DOI: 10.1038/s41586-022-05260-5
Researchers at the Technion—Israel Institute of Technology have developed a technique to measure the long-term effects of antibiotic combinations, or cocktails. These combinations are of serious interest to the scientific and medical communities because the use of single antibiotics often leads to the rapid development of bacterial resistance to these drugs.
The research published in Nature was led by Technion researchers Professor Roy Kishony from the Faculty of Biology and Dr. Viktória Lázár, a postdoctoral student in his lab who is now working at the Synthetic and Systems Biology Unit of the Biological Research Centre in Szeged, Hungary.
The researchers discovered that in many cases, a combination of several antibiotics may actually reduce the treatment's effectiveness in the long term—meaning that the combination of drugs could prove to be inferior to the success of each individual drug. However, they point to specific combinations that do manage to prevent the development of resistance and thus protect the patient for a long period from the aggressive bacteria.
The bacterium tested in the study is Staphylococcus aureus, a particularly violent bacterium that has developed resistance to many types of antibiotics. This bacterium is responsible for a significant part of nosocomial (in-hospital or in-clinic) infections. The study was conducted both in cultures of this bacterium in the lab and in an animal model—larvae of the Galleria mellonella moth.
Antibiotics are a family of drugs that play a central role in modern medicine and save lives on a daily basis. The natural antibiotic substances that developed during evolution in fungi and yeast were discovered about a century ago in the research of London's Sir Alexander Fleming, Australia-born Howard Walter Florey, and immigrant to England from Berlin who was of Russian-German-Jewish descent Ernst Boris Chain. The three shared the Nobel Prize in Physiology or Medicine for 1945.
In the past century, antibiotic treatment has saved hundreds of millions of people.
However, the success of antibiotic therapy has turned into a double-edged sword because the widespread use of these anti-bacterial drugs leads to the evolutionary development of bacteria that develop resistance. This trend raises a justified fear of a post-antibiotic era, or a period when bacteria will no longer respond to antibiotic drugs and people will die, as in the past, from infections that are now considered mild and not dangerous.
The laboratory of Prof. Kishony, one of the leading experts in the field of antibiotic resistance, develops methods that make it possible to estimate in advance the resistance of a given bacterium to a given antibiotic in the present and even to predict the resistance level it is expected to develop in the future. In the current study, a combination of different antibiotic drugs that prevent the formation of resistance was examined.
The researchers noted that the COVID-19 pandemic has increased the use of antibiotics, even though SARS-CoV-2 is not affected by antibiotics because it is a virus and not a bacterium. However, giving antibiotics helps COVID-19 patients to avoid secondary bacterial infections. With the growth of antibiotic use, the evolution of resistant Staphylococcus aureus strains accelerates.
To summarize, the Technion researchers discovered that combinations of antibiotics may harm the effectiveness of the treatment and point to specific combinations that speed up or inhibit the development of resistant bacteria. In doing so, they help pave the way for more effective treatments and the curbing of the "resistance epidemic" that threatens humanity.
More information: Viktória Lázár et al, Antibiotic combinations reduce Staphylococcus aureus clearance, Nature (2022). DOI: 10.1038/s41586-022-05260-5
Journal information: Nature
Provided by Technion - Israel Institute of Technology