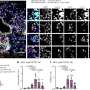
Fig 1. Genome-wide associations of AD. A, A Manhattan plot of AD in 796,661 Europeans. The pink dashed line corresponds to P = 5 × 10−8. The novel loci are highlighted with pink font. B, Functional annotation of the significant variants and variants in LD with significant variants (see this article’s Online Repository at www.jacionline.org). Bars are colored by -log2(enrichment) relative to all variants in the reference panel. C, Significant (P < 6.36 × 10−6) gene sets obtained using MAGMA (see this article’s Online Repository at www.jacionline.org). D, Population-specific effect sizes and corresponding 95% CIs of the lead variants in the 30 reported loci. E, Association results of the lead variants in the 30 reported loci in FinnGen using 3 different AD case definitions (see this article’s Online Repository at www.jacionline.org). “AD” cases had an entry of ICD-10 code L20 or ICD-8 code 691, and individuals with no record of these were considered as controls; participants with ICD-9 code 6918X were excluded from the analyses. “AD reimb” cases had an entry of KELA reimbursement codes 134 (erythrodermia exfoliativa universalis), 395 (dupilumab), or 317 (topical calcineurin inhibitors). “AD strict” cases had an entry of ICD-10 code L20.0, ICD-9 code 6918B, or ICD-8 code 69100. In Fig 1, D and E, novel variants are highlighted with bold black font. Credit: DOI: 10.1016/j.jaci.2021.07.043
An international study led by the University of Oulu's researchers reports five new genetic loci predisposing to atopic dermatitis. Based on three extensive biobanks, the study helps to understand the pathogenic mechanisms behind the disease and may open up opportunities for developing new forms of treatment.
Atopic dermatitis (atopic eczema) is an inflammatory skin disease affecting approximately 10–20% of the population. Genetic factors are known to contribute to the development of the disease, and around twenty genes associated with atopic dermatitis are already known.
The five loci now identified function as part of biological signaling pathways that have not previously been associated with atopic dermatitis.
According to the researchers, some of the most interesting findings were the new atopic dermatitis risk associations near DSC1 and SERPINB7—these genes produce proteins that enhance the strength of the skin's surface. In addition, the researchers identified a locus in the vicinity of the IL22 and IFNG genes that likely influences the risk of atopic dermatitis by modulating the immune system.
The genome-wide analysis made use of biobanks in Finland, Estonia and the United Kingdom. In total, the genome and health data of nearly 800,000 people were analyzed. The Finnish research material is part of the FinnGen project.
"Atopic dermatitis is a common disease, and new ways of treating it are needed. More detailed research into the genetic loci we have now identified may lead to the development of novel therapies in the longer term. In addition, as more biobank data are collected, we may be able to identify more new mechanisms behind atopic dermatitis, as well as rarer, population-specific genetic variants predisposing to the disease," says Anu Pasanen, Postdoctoral Researcher at the University of Oulu.
"Our results show that by using biobank-based genetic and register data, we can get valuable information about the molecular mechanisms contributing to disease susceptibility. Similar studies could also be utilized more in pharmaceutical development," says Eeva Sliz, Postdoctoral Researcher at the University of Oulu.
The study was published in the Journal of Allergy and Clinical Immunology.
More information: Eeva Sliz et al, Uniting biobank resources reveals novel genetic pathways modulating susceptibility for atopic dermatitis, Journal of Allergy and Clinical Immunology (2021). DOI: 10.1016/j.jaci.2021.07.043
Journal information: Journal of Allergy and Clinical Immunology
Provided by University of Oulu