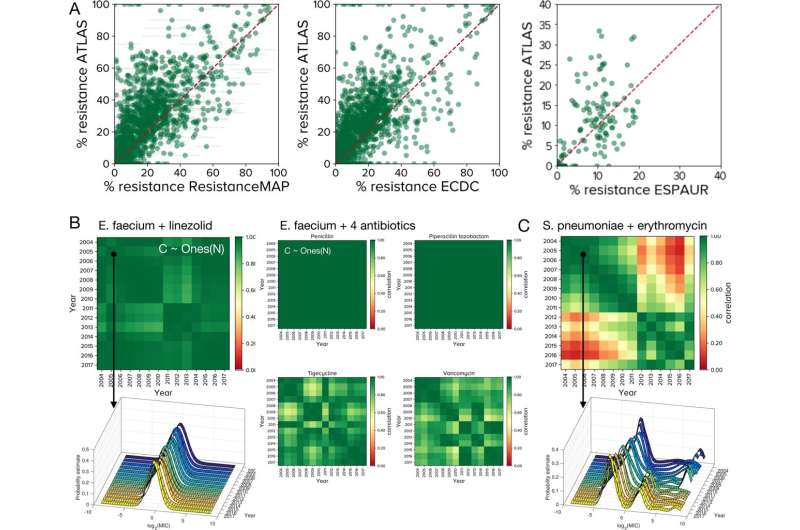
Between database comparisons with ATLAS. A Each point represents a PA (pathogen-antibiotic) pair in a given country, for a given year with frequency of resistance (fR as a %age) on x and y axes. ATLAS tends to over-estimates fR relative to ResistanceMap, ECDC and ESPAUR data: differences between fR in ATLAS and other databases are positively skewed, however between-database differences are smaller for larger PA pair datasets (Supplementary Fig. 2 has statistics). B Between-year correlations for many ATLAS PA pairs form correlelograms, called `C' here, that are close to the (pure green) unity matrix of ones, Ones(N), for N years. 5 PA pairs for Enterococcus faecium are shown. (Supplementary Fig. 8C has some correlelogram statistics, Supplementary Fig. 9 shows many are close to Ones(N) but not all, see Fig. 3 and Supplementary Fig. 10). The left column shows the global MIC (minimal inhibitory concentration) distribution of E. faecium and linezolid is stable from year to year and its correlelogram is close to Ones(N). The middle panel shows 4 correlelograms with banded structures that occur when MIC distributions experience change. C This correlelogram of Streptococcus pneumoniae and erythromycin have a block structure because their MIC distributions correlate poorly between years: a high-MIC cluster diminishes and is replaced by a cluster with lower MIC in 2010–2011 (c.f. Fig. 4). Credit: Nature Communications (2022). DOI: 10.1038/s41467-022-30635-7
Scientists have used ideas from artificial intelligence to identify patterns of antibiotic resistance around the world.
An international team of scientists, including Professor Robert Beardmore of the University of Exeter's Living Systems Institute, used mathematical modeling to extract antibiotic resistance patterns from 6.5 million datapoints collected worldwide.
The researchers examined a host of datasets from medical companies and health agencies to identify patterns in how antibiotic resistance changes.
The most medically complete dataset is published by Pfizer—which has been collating data for two decades.
Using this dataset, called ATLAS, the team were able to compare information already in the public domain to highlight discrepancies and predict future increases in resistance.
Authors found that ATLAS provides a significantly different picture, including resistance not seen previously. They also highlight the lack of data made available in Africa.
The researchers believe the new techniques will provide a significant boost to both the understanding and knowledge of how resistance is growing, and where.
The research was published in Nature Communications on Wednesday May 25, 2022.
Professor Beardmore, lead author of the study said: "AI is a box of tricks that could help solve the antibiotic resistance problem, but national health agencies need to publish much more data for that to happen.
"Resistance does seem to be on the increase but even if it were to come down because of successful health policy changes or new medical technologies, missing data make those reductions hard to spot."
Antibiotic resistance is recognized as being one of the most significant threats to global health. Antibiotic resistance can affect anyone, of any age, in any country.
While it occurs naturally, the overuse and misuse of antibiotics in humans is causing the process to accelerate at a significant rate. As a result, a growing number of illnesses, such as sepsis, pneumonia and tuberculosis, are becoming harder to treat.
A key factor in the fight against resistance lies in the patterns that highlight where outbreaks might occur, and for what ailments. These new techniques provide a pivotal tool for this, finding new ways of extracting antibiotic resistance patterns from many millions of datapoints.
Pablo Catalan, a data scientist from Madrid said: "Our work on ATLAS shows resistance in patients is a dynamic process that contains lots of patterns and the more data we have, the better we'll understand those patterns and understand when our actions are pulling resistance downwards, or making things worse."
Jon Iredell, a collaborating clinician added: "When a bacterium is sampled from one of my patients, that is a chance to increase the amount of data we have on patterns of antibiotic resistance that we are not, as yet, taking."
"Seeking patterns of antibiotic resistance in ATLAS, an open, raw MIC database with patient metadata" is published in Nature Communications.
More information: Pablo Catalán et al, Seeking patterns of antibiotic resistance in ATLAS, an open, raw MIC database with patient metadata, Nature Communications (2022). DOI: 10.1038/s41467-022-30635-7
Journal information: Nature Communications
Provided by University of Exeter