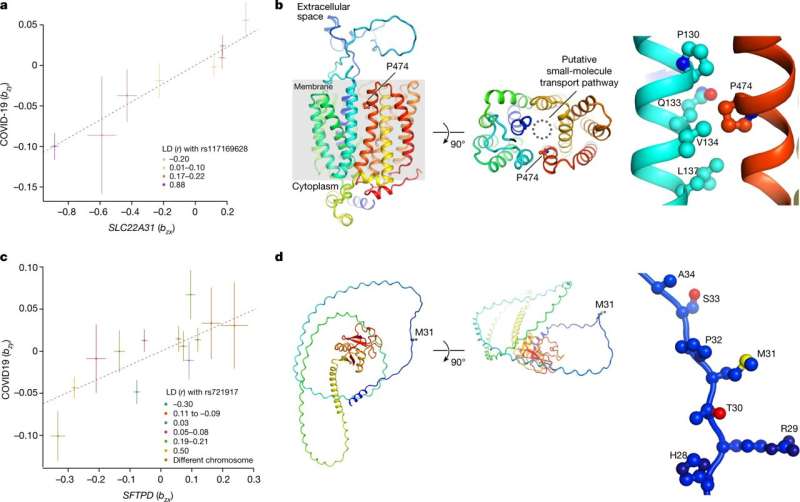
Functional genomics analyses for SLC22A31 and SFTPD. a, Effect-size plot for the effect of multiple variants on SLC22A31 expression (eQTLgen, x axis) against increasing susceptibility to critical COVID-19 (βxy = 0.11; Pxy = 1.3 × 10−9). The color shows linkage disequilibrium (LD) with the missense variant rs117169628. b, Three cartoon views of an AlphaFold model of putative solute carrier family 22 member 31 (SLC22A31; UniProtKB: A6NKX4). The side chains of Pro474 and interacting amino acids are shown as connected spheres. A putative channel for small-molecule transport across the cell membrane is indicated by a dashed circle. Pro474 is predicted to be located in the transmembrane helix and point towards a putative transport pathway of a small molecule. The risk variant, P474L (Ala at rs117169628) would be expected to introduce more flexibility to the transmembrane helix and might therefore affect the transport properties of SLC22A31. Pro474 is predicted to be in a tightly packed environment, and may therefore affect the folding of SLC22A31. c, Effect-size plot for effect of multiple variants on SFTPD expression (eQTLgen, x axis) against increasing susceptibility to critical COVID-19 (βxy = 0.16; Pxy = 9.7 × 10−6). Color shows linkage disequilibrium with the missense variant rs721917. d, Three cartoon views of an AlphaFold model of pulmonary surfactant-associated protein D (SFTPD; UniProtKB: P35247). The side chain of the variant Met31 is shown as connected spheres. Met31 is predicted to be located in the secondary-structure-lacking region of SFTPD. In the diagram on the right, oxygen and nitrogen atoms are colored red and blue respectively, and the sulfur atom is colored yellow. Credit: Nature (2023). DOI: 10.1038/s41586-023-06034-3
Researchers at the University of Edinburgh in the U.K. have led a study in collaboration with scientists worldwide, looking into cases of critical illness in COVID-19 patients.
Critical illness in COVID-19 is an extreme and clinically consistent disease phenotype the team has found presenting in patients with shared genetic attributes. These shared genetics hint at a shared mechanism for the critical illness not seen in other patients and potential therapies to address the condition.
Patients with confirmed COVID-19 and requiring continuous cardiorespiratory monitoring or organ support (a generalizable definition for critical illness) were recruited in 2020–2022.
Researchers analyzed 24,202 cases of COVID-19 with critical illness with a combination of microarray genotyping and whole-genome sequencing data from the international GenOMICC study (11,440 cases) and other studies recruiting hospitalized patients with severe and critical illness, including the COVID-19 Human Genetics Initiative, the International Severe Acute Respiratory and Emerging Infection Consortium, the Spanish Coalition to Unlock Research on Host Genetics consortium and 23andMe.
The team found 49 genome-wide significant associations, of which 16 have not been reported previously and 196 significantly associated genes in a gene-level analysis. Although the implicated variants are not directly causing illness in the patients, they can highlight molecular mechanisms that make some COVID-19 infections much more severe. The findings are published in the journal Nature.
Many genes implicated in critical COVID-19 are highly expressed in the monocyte-macrophage system, which has poor coverage in existing expression quantitative trait loci datasets. Macrophages synthesize many substances involved in host defense and inflammation and play a pivotal role in immune system reactions.
Additionally, the investigation found variation in circulating protein levels with 15 unique proteins linked to critical illness and some with well-studied biomarkers that make them good candidates for drug targeting.
The research has identified several potential druggable targets in multiple systems, including inflammatory signaling, monocyte-macrophage activation and endothelial permeability. Some of the targets found have already seen positive results with therapeutic signals in multiple drug trials, providing a good proof-of-concept for drug target identification using comparative genetics.
More information: Erola Pairo-Castineira et al, GWAS and meta-analysis identifies 49 genetic variants underlying critical COVID-19, Nature (2023). DOI: 10.1038/s41586-023-06034-3
Journal information: Nature
© 2023 Science X Network