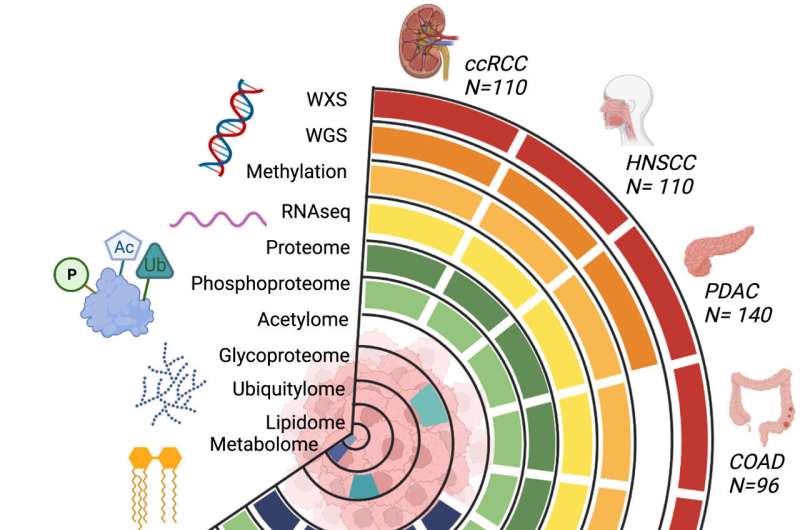
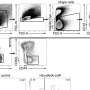
Tumor types and data types of the CPTAC pan-cancer dataset. Overview of the available molecular data types for the CPTAC pan-cancer cohort (n = 1072, see Table S1 for list of excluded cases and reasons for exclusion from the original datasets). Whole exome, whole genome, methylation, transcriptome, proteome, and phosphoproteome data are available for all ten cancer types. Normal samples are available for a subset of tumor types, see Tables S1 and S2. Credit: Cancer Cell (2023). DOI: 10.1016/j.ccell.2023.06.009
The National Institutes of Health is releasing a comprehensive dataset that standardizes genomic, proteomic, imaging, and clinical data from individual studies of more than 1,000 tumors across 10 cancer types. Researchers from around the world will be able to use this publicly available resource to uncover new molecular insights into how cancers develop and progress. The dataset was generated by the Clinical Proteomic Tumor Analysis Consortium (CPTAC) at the National Cancer Institute, part of the National Institutes of Health.
The pan-cancer proteogenomic dataset, which is described in a paper published in Cancer Cell, builds on decades of technological advances in proteomic science. The launch of this dataset supports the Biden-Harris Administration's 'Cancer Moonshot' goal of accelerating cancer research through improved sharing of data.
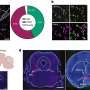
Two additional research papers published in Cell by CPTAC investigators provide an initial demonstration of the dataset's potential as a valuable resource for scientific discovery. In the first paper, multi-omic analyses are used to link cancer driver mutations with protein patterns. The second paper delves into protein modifications that regulate cell signaling and physiology to show associations with DNA repair, metabolism, and immunity across different tumor types.
The pan-cancer proteogenomic dataset will be publicly available through the NCI Cancer Research Data Commons repositories. Proteomics data can be accessed via the Proteomic Data Commons. Genomic and transcriptomic data can be accessed via the Genomic Data Commons and the Cancer Data Service.
More information: Yize Li et al, Proteogenomic data and resources for pan-cancer analysis, Cancer Cell (2023). DOI: 10.1016/j.ccell.2023.06.009
Yize Li et al, Pan-cancer proteogenomics connects oncogenic drivers to functional states, Cell (2023). DOI: 10.1016/j.cell.2023.07.014
Yifat Geffen et al, Pan-cancer analysis of post-translational modifications reveals shared patterns of protein regulation, Cell (2023). DOI: 10.1016/j.cell.2023.07.013
Journal information: Cell , Cancer Cell
Provided by National Institutes of Health