Colorectal cancer develops in what is probably the most complex environment in the human body, a place where human cells cohabitate with a colony of approximately 10 trillion bacteria, most of which are unknown. At the 2014 American Association for Cancer Research Annual Meeting in San Diego, researchers from The Wistar Institute will present findings that suggest the colon "microbiome" of gut bacteria can change the tumor microenvironment in a way that promotes the growth and spread of tumors.
Their results suggest that bacterial virulence proteins may suppress DNA repair proteins within the epithelial cells that line the colon. The research opens the possibility of modifying colon cancer risk by altering the population makeup of bacteria in the intestines of people at risk due to genetics or environmental exposure.
"There is a drastic, unmet need to look at new ways to define exactly how colon cancer forms in the gut and what triggers its progression into a lethal form," said Frank Rauscher, III, Ph.D., a professor in The Wistar Institute Cancer Center. "We suggest that some bacterial proteins can promote genetic changes that create conditions in the gut that would favor the progression of colon cancer."
While colorectal cancer incidence rates have declined, likely due to more widespread screening, survival rates have not. According to the American Cancer Society, about 50,000 Americans will die from colorectal cancer this year. "While our understanding of the gene mutations involved in colon cancer has improved, this has not lead to the promised increases in overall survival," Rauscher said.
Intestinal bacteria typically provide many benefits to their human hosts, aiding in digestion and crowding out more directly pathogenic bacteria. However, both "friendly" commensal bacteria and infective, pathogenic bacteria have been shown to actively reduce inflammation, an important tool used by the human innate immune system to promote healing and prevent the spread of infection.
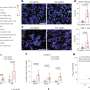
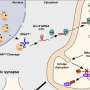
In these studies, Rauscher and his colleagues injected anti-inflammatory proteins produced by EPEC (Enteropathogenic Escherichia coli) bacteria into colon epithelial cells. One of these proteins, NLEE, is an enzyme that targets TAB2, a human scaffolding protein involved in the transduction of chemical signals in the NF-κB pathway. Targeting TAB2 results in the inactivation of numerous inflammatory activities in the gut.
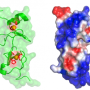
Rauscher and colleagues looked for other human proteins that could be targeted by NLEE. Remarkably, they found that NLEE also has the capability of shutting off a protein, ZRANB3 involved in DNA repair. If bacterially infected colon cells can no longer repair damage to their DNA, mutations will accumulate, which will promote cancer growth.
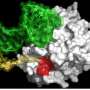
In addition, along with collaborators in the laboratory of Feng Shao, Ph.D., at the National Institute of Biological Sciences in Beijing, China, they demonstrated that NLEE proteins attack TAB2 and ZRANB3 by methlylating these proteins—essentially adding a single methyl molecule—which unfolds the target proteins. NLEE appear to specifically attack a structure on TAB2 and ZRANB3 known as a "zinc finger," which is a common structural motif used in many other proteins. When the researchers determined the structure of NLEE, they found a deep cleft on the protein specific to a certain zinc finger pattern. A survey of EPEC-infected colon cells showed that this zinc finger pattern was common to at least three DNA repair enzymes, suggesting that NLEE has the capability of having widespread influence on mechanisms in the colon that typically prevent cancer growth.
"Our results suggest that some infective intestinal bacteria, which normally can simply cause gastric distress, have the capability of inducing genetic changes (by limiting repair) in our intestinal cells which could lead to tumor development," Rauscher explained. "It is possible that limiting the amount of this bacteria in our gut may protect us from the genetic changes which accumulate in our intestinal cells over time and lead to cancer development."
According to Rauscher, this is a new way to look at the microenvironment in the gut as an incubator for colon cancer, depending upon which type and species of bacteria are resident and potentially infectious in our large intestines. Rauscher and his collaborators are currently embarking on a project to further test their hypothesis.
More information:
Presentation Title: Control of DNA repair and genome stability by the colon microbiome: The EPEC bacterially encoded NLEE virulence effector protein methylates and inactivates the human ZRANB3 DNA repair helicase
Date: Tuesday, April 8, 2014, 4:35 PM - 4:50 PM
www.abstractsonline.com/Plan/V … 3-29ab3d1e6bd0&mKey=%7b6FFE1446-A164-476A-92E7-C26446874D93%7d
Provided by The Wistar Institute