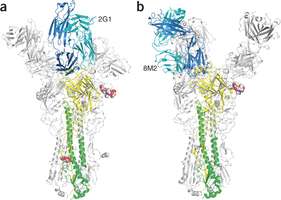
Structures of H2 HA–VH1-69 antibody complexes. Credit: Nature Structural & Molecular Biology, doi:10.1038/nsmb.2500
(Medical Xpress)—Researchers with the Scripps Institute have discovered that three naturally occurring antibodies are able to overcome flu mutations by attaching to a non-changing protein in the flu virus. As they describe in their paper published in the journal Nature Structural & Molecular Biology, they have found that the antibodies that are able to defeat the flu mutations can also stop an infection.
In order for a flu virus to infect a person, it must attach itself to cells in the respiratory tract. To do that, it uses a protein called hemagglutinin to attach to sugars in human cells called sialic acid. In this new research, the team found that hemagglutinin has a cavity shape, and that some naturally occurring antibodies are able to move into the cavity, preventing the virus from attaching to the cell, and thus an infection. More importantly, they have found that even as the flu virus mutates—to create shapes that fool antibodies sent by the body to destroy them—the cavity's size and shape remains the same. This means, that if a way could be found to exploit the same weakness found by some antibodies, a universal vaccine could be created that would work against all types of the flu viruses, instead of just one strain.
A flu virus infection is actually a little more complicated than that—the virus doesn't just attach to one part of the cell via sialic acid, it attaches to many. Multiple attachments make it almost impossible for antibodies to work against them. The researches refer to it as similar to Velcro. This is why current treatments don't work as well as hoped—their mission is to assist antibodies in removing flu viruses already attached to cells.
Making things even more difficult for the person trying to fight the infection, is that the flu viruses also multiply, using mutations to thwart invading antibodies. A better approach to fighting such an infection, the researchers say, would be to create a substance similar to an antibody that could insert itself between a flu virus (in its cavity) and the cell it's trying to infect, preventing attachment. Doing so would make moot any mutations carried out by the flu virus, as the cavity remains the same in all variations. The end result would be the prevention of an infection.
This new research shows what needs to be done to conquer the flu virus once and for all—the next step will involve trying to figure out how to create an artificial antibody, or to learn how to coax natural ones into performing as desired.
More information: A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin, Nature Structural & Molecular Biology (2013) doi:10.1038/nsmb.2500
Abstract
Influenza virus hemagglutinin (HA) mediates receptor binding and viral entry during influenza infection. The development of receptor analogs as viral-entry blockers has not been successful, which suggests that sialic acid may not be an ideal scaffold to obtain broad, potent HA inhibitors. Here, we report crystal structures of Fab fragments from three human antibodies that neutralize the 1957 pandemic H2N2 influenza virus in complex with H2 HA. All three antibodies use an aromatic residue to plug a conserved cavity in the HA receptor-binding site. Each antibody interacts with the absolutely conserved HA1 Trp153 at the cavity base through π-π stacking with the signature Phe54 of two VH1-69–encoded antibodies or a tyrosine from HCDR3 in the other antibody. This highly conserved interaction can be used as a starting point to design inhibitors targeting this conserved hydrophobic pocket in influenza viruses.
Journal information: Nature Structural & Molecular Biology
© 2013 Medical Xpress