Prostate cancer is one of the most common cancers in men. Tumor growth is critically regulated by the androgen receptor, and treatment strategies to lower androgens, such as testosterone, are a mainstay of clinical treatment. Over time, however, resistance frequently develops and the disease may progress to a castration-resistant form that expresses a variant of the androgen receptor, which evades blockade by drugs that dampen androgen receptor signaling. Thus, clinically there is an urgent need to identify patients expressing androgen receptor variants to determine which treatment options are likely to benefit patients.
In this issue of JCI Insight , researchers at the University of British Columbia describe a new imaging tool to detect the presence of the androgen receptor and its active splice variants.
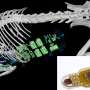
Led by Marianne Sadar, the research group developed an analog of an investigational drug that binds to portions of the androgen receptor that are common to the full length and variant forms of the androgen receptor.
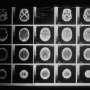
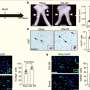
In a mouse model of prostate cancer, they showed that this compound specifically detected prostate cancer cells expressing androgen receptor by SPECT/CT imaging.
These findings suggest that this imaging agent may be suitable for further development for castration-resistant prostate cancer patients.
More information: Yusuke Imamura et al, An imaging agent to detect androgen receptor and its active splice variants in prostate cancer, JCI Insight (2016). DOI: 10.1172/jci.insight.87850
Provided by JCI Journals