Credit: Agency for Science, Technology and Research (A*STAR), Singapore
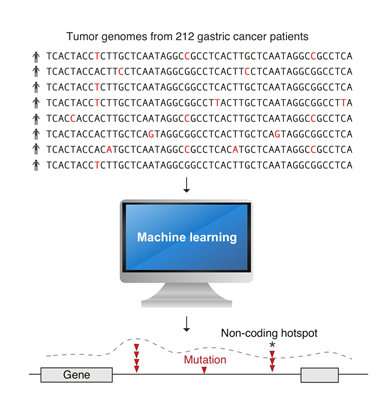
In a pioneering study, scientists at A*STAR's Genome Institute of Singapore (GIS) have developed new machine learning computer models, a type of artificial intelligence (AI), to accurately pinpoint cancer mutations. They have also discovered new mutations in non-coding DNA (specifically, DNA that does not encode for proteins) which may cause gastric cancer. Furthermore, the innovative methods and technology developed through this study will aid researchers in understanding the impact of mutations in non-coding DNA in other cancer types.
Cancer is one of the leading causes of death worldwide and gastric cancer (also known as stomach cancer) is the fourth most lethal cancer3 in the world. It arises from mutations in the DNA that lead to abnormal cell growth. Much has been learnt about cancer through the study of two percent of DNA that comprise our genes. However, the other 98%, termed non-coding DNA, is still mostly unchartered territory. Non-coding DNA regulates activity of the genes and there is increasing evidence that mutations in these regions can also contribute to cancer.
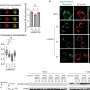
In this project, two new AI methods were created to scan the entire genomes of 212 gastric cancer tumours in a few months. The analysis would have otherwise taken 30 years to complete on a standard modern computer. Using computer clusters at GIS and the National Supercomputing Centre (NSCC) Singapore, the analysis uncovered several new cancer-associated mutation hotspots located throughout the genome. It also provided new evidence that mutations in the non-coding DNA may cause cancer by altering the 3-dimensional (3D) genome structure.
Dr Anders Skanderup, Principal Investigator at GIS and lead scientist of the study, said, "We focus on computational and data-driven approaches to study the root of cancer so as to develop better strategies to combat it. Our findings suggest that mutations at 11 non-coding sites regulating the 3D genome structure are staggeringly frequent. Approximately one in every four gastric cancer patients have mutations at these specific sites."
He continues, "These non-coding mutations are also frequent in other types of gastrointestinal cancers such as colorectal, pancreatic and liver cancer. Therefore, they can be used as biomarkers to detect and monitor the progression of gastrointestinal cancers too."
Professor Patrick Tan, Director of Singhealth Duke-NUS Institute of Precision Medicine (PRISM), Deputy Executive Director of A*STAR's Biomedical Research Council, and co-lead scientist said, "Sophisticated machine learning techniques such as the one developed in this study are absolutely essential towards decoding the information encoded in our genomes. If experimentally validated, these findings point towards a mechanism of cancer development missed by previous studies." Professor Tan is also a professor at the Cancer & Stem Cell Biology Programme in Duke-NUS Medical School.
Professor Ng Huck Hui, Executive Director of GIS, said, "Previous studies focus solely on profiling mutations in the protein coding regions of our DNA, which makes up a mere two percent of our DNA. So, it has been an open question for ages whether we are missing vital information by overlooking the other vast 98%. This is the first study investigating the impact of non-coding DNA mutations in gastric cancer and we anticipate that it will inspire new research to further uncover the mechanisms and impact of these specific mutations."
More information: Yu Amanda Guo et al. Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers, Nature Communications (2018). DOI: 10.1038/s41467-018-03828-2
Journal information: Nature Communications