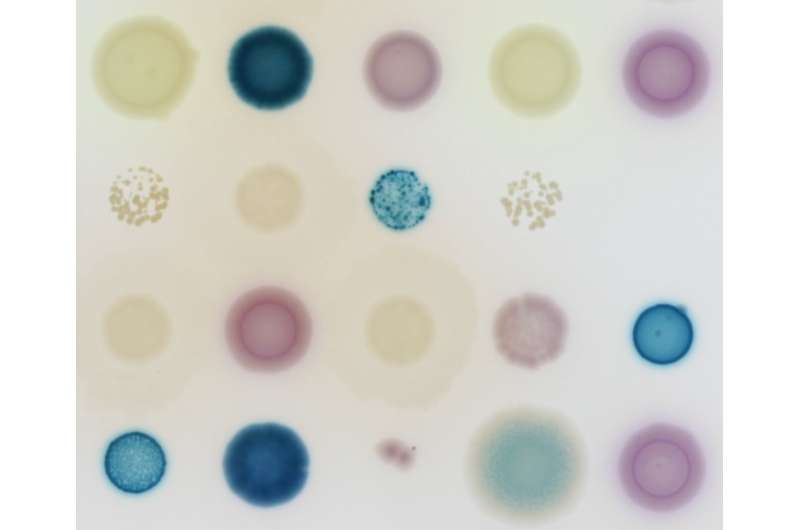
Assistant professor Dr. Marjon de Vos, last author of the review paper in Lancet Infectious Diseases. Credit: Wageningen University and Research
Taking an evolutionary view can inspire new ideas in clinical microbiology. For example, evolutionary studies can reveal why some antimicrobial dosing regimens are better than others in preventing the development of drug resistance. Looking at microbial communities, rather than just pathogenic micro-organisms, can also lead to new insights. That is why clinicians, bioinformaticians analysing pathogens, and evolutionary biologists should all work together. These are the conclusions of a diverse group of scientists led by University of Groningen microbiologist Marjon de Vos, in a short review published by The Lancet Infectious Diseases on 30 April.
As a microbiologist, De Vos studies urinary tract infections. She realized that a lot could be gained by collaborating with different specialists, such as clinical microbiologists, research scientists working in evolution and ecology, and bioinformaticians. "For example, I noticed that the bacteria involved communicate with each other and can form a stable ecosystem, which affects their susceptibility to antibiotics." This realization led to an interdisciplinary workshop in 2017, which in turn resulted in the review paper now published in The Lancet Infectious Diseases.
Cystic fibrosis
"The main question in the workshop and the paper is how we can use knowledge from evolutionary theory and bioinformatics to solve clinical problems," says De Vos. Bioinformatics is needed to analyse the vast amount of genetic data collected on infectious diseases. Evolutionary theory can help to explain the observed patterns in adaptation and predict how treatments will affect the ecosystem in which pathogens live.
De Vos: "An example that we discuss in the paper is the use of cyclic antibiotic treatments in cystic fibrosis patients." These patients suffer from chronic lung infections, for which they receive antibiotics. In order to minimize the development of drug resistance, they can potentially be treated alternately with two different drugs. The idea is that pathogens that become resistant to one drug will still be susceptible to the other. "This has led to positive results. However, there is a danger that this treatment will induce multi-drug resistance, so it is important to check the response of the pathogens in the lab first in order to find the optimal treatment schedule."
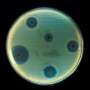
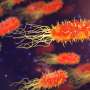
Bacteria isolated from urinary tract infections, grown on diagnostic agar which gives each bacterial species its own colour. Credit: Marjon de Vos
Plasmids
Analysis of the DNA of pathogens has produced a lot of new information on the genetic basis of antibiotic resistance. "Yet, we still don't know how changes in genes lead to the different characteristics of these pathogens. We need experiments by evolutionary biologists in order to understand the link between the genotype, the DNA sequence and the phenotype—for instance, the level of resistance."
Another example is the spread of antibiotic resistance genes. These are often present on plasmids, i.e. bits of circular DNA that can easily be exchanged between bacteria. "In this case, it is important to note that bacteria often have to adapt to carrying a new plasmid. If a bacterium acquires a plasmid to which it is not adapted, this could slow its growth rate considerably. In that case, there is a trade-off between resistance on the one hand and the fitness and growth rate of bacterium on the other, which means a resistance gene on the plasmid may not have much of a clinical effect. An ecological or evolutionary approach may aim to exploit such trade-offs to limit the spread of antimicrobial resistance." The question in monitoring antibiotic resistance, according to De Vos, shouldn't only be where it occurs but how it arises in bacterial communities. "To understand what is causing the selection and propagation of these genes, we need a much wider perspective on these processes."
Adaptation
Collaboration between different specialists requires the investment of all involved. "We really need to engage with each other, as we all tend to look at different aspects of infections. Only by discussing these aspects together is it possible to see when an observation from evolutionary theory is relevant to clinical practice." "A final example shows how this can happen," says De Vos: "Sandra Breum Andersen, the lead author of the paper, found that bacteria that cause chronic lung infections in cystic fibrosis patients often evolved in a certain way that was assumed to be due to an adaptation to the host. However, laboratory experiments showed that this was not due to host adaptation but, instead, to the bacteria reacting to each other by stealing each other's resources. Thus, instead of focusing on the host-bacteria interaction of the disease, meddling in the social lives of these bacteria could potentially lead to novel treatments."
More information: Sandra B Andersen, B Jesse Shapiro, Christina Vandenbroucke-Grauls, Marjon G J de Vos: Microbial evolutionary medicine: from theory to clinical practice. The Lancet Infectious Diseases (2019). DOI: 10.1016/S1473-3099(19)30045-3
Journal information: Lancet Infectious Diseases
Provided by University of Groningen