In March 2020, the World Health Organization declared a global COVID-19 pandemic and the UK announced a strict national lockdown. When Oxford University scientists were sent home they switched on their computers and started to help develop new drugs to target SARS-CoV-2, the virus that causes COVID-19.
An international collaboration, involving 29 scientists from around the world, focused on understanding how SARS-COV-2 makes its worker proteins at the molecular level so we could develop novel antiviral drugs and block their production—all working from home but coming together weekly on Zoom to tackle this terrifying disease.
'If scientists can design new molecules that bind more tightly than these natural substrates, they could stop the virus dead in its tracks. Blocking the cutter stops the virus from replicating—a strategy that has worked for treating other viral diseases like HIV and hepatitis.' Said Prof. Chris Schofield, Profess or of Chemistry, Oxford University
Despite the development of successful vaccines in record time, there are no drugs that have been designed specifically to target COVID-19, but they are desperately needed.
Once SARS-CoV-2 has invaded a healthy human cell, the virus's own genetic material commandeers the infected cell's machinery, forcing it to make new copies of the virus.
The new virus begins as one long protein that cuts itself into functional units. First the cutting catalyst or 'protease' cuts itself out, which then cuts at multiple other positions.
SARS-CoV-2 has two molecular machines or proteases that resemble 'molecular scissors'. One of these, called the main protease, or 'Mpro' for short, cuts at no less than 11 of these cut sites.
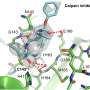
Using 3D structures obtained by shining X-rays onto crystals of the main protease of SARS-CoV and SARS-CoV-2, Prof. Morris and his collaborators were able to develop computational models of how the SARS-CoV-2 Mpro binds to its 11 cut sites. From these models, they were able to gain key insights into how these viral Mpro 'molecular scissors' work.
'What is remarkable about this collaboration, involving 29 scientists from around world, is that every meeting was entirely virtual, with many collaborators yet to meet face-to-face.' Said Prof. Garrett M. Morris from Oxford University.
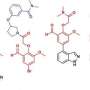
Building on this knowledge, and using computational methods, they next sought to design novel molecules that could bind even more tightly than the natural cut sites. Using computers to sift through just over 200 trillion possibilities, they proposed new molecules that would stop the virus from maturing.
All 11 cut sites and 4 of these designed peptides were synthesized and tested in the laboratory of Prof. Chris Schofield in the Chemistry Research Laboratory at the University of Oxford. The experiments showed that the novel designed peptides not only bound to the molecular scissors but blocked the substrates and actually inhibited the Mpro.
Scientists also carried out an extensive analysis of hundreds of published 3D structures of small molecules bound to Mpro and predicted how inhibitors designed by the COVID Moonshot would bind, figuring out how these 'molecular keys' fitted into the 'molecular lock' of Mpro, and using these insights to propose how to design new drugs to treat COVID-19.
A vast array of computational techniques was employed to build a complete picture of SARS-CoV-2 Mpro, ranging from comparative molecular modelling, molecular dynamics, interactive molecular dynamics in virtual reality, quantum mechanics, computational peptide design, protein-ligand docking, protein-peptide docking, and protein-ligand interaction analysis.
More information: H. T. Henry Chan et al, Discovery of SARS-CoV-2 Mpro Peptide Inhibitors from Modelling Substrate and Ligand Binding, Chemical Science (2021). DOI: 10.1039/D1SC03628A
Journal information: Chemical Science
Provided by University of Oxford