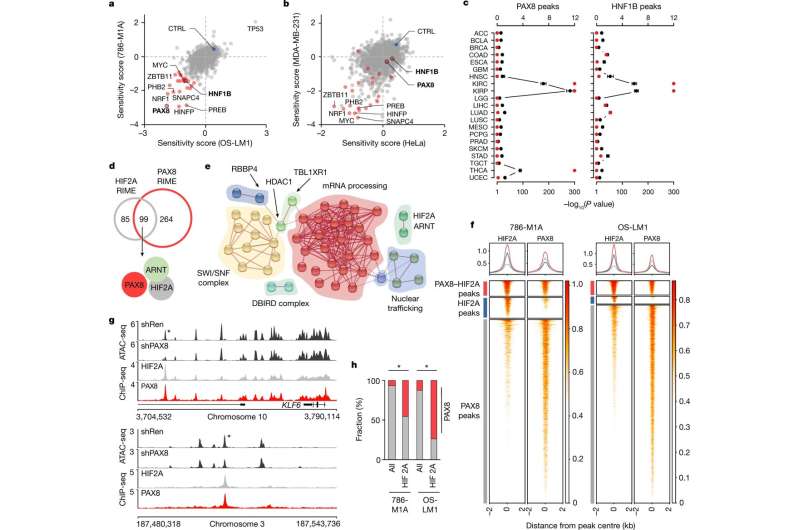
Chromatin level interaction between the renal lineage factor PAX8 and oncogenic HIF2A in ccRCC. a,b, Pooled CRISPR–Cas9 loss-of-function screen results of ccRCC cell lines (a) and non-ccRCC cell lines (b). Sensitivity score, log2 of the mean of the top three depleted sgRNAs per gene, two replicates per condition, at the end of the assay compared with the start of the assay. ccRCC dependencies are in red. CTRL, average of non-targeting controls. c, Overlap between cancer-type-specific ATAC-seq peaks in TCGA data and those with reduced accessibility after PAX8 and HNF1B depletion in ccRCC cells. Top axis, odds ratio of overlap (black), 95% confidence interval. Bottom axis, P value, one-sided Fisher’s exact test (red). d, Overlap between PAX8- and HIF2A-interacting proteins as determined by RIME in 786-M1A cells. e, Network presentation of physical connections between 89 shared nuclear proteins from HIF2A and PAX8 interactomes. Protein names are provided in Extended Data Fig. 4a. f, Heatmaps of HIF2A and PAX8 ChIP-seq signals from 786-M1A and OS-LM1 xenografts (three tumors each) across regions with strong PAX8–HIF2A co-binding (red), predominant HIF2A binding (blue) and predominant PAX8 binding (gray). Top panels show the average signal within each of the three categories in the same colors. g, HIF2A and PAX8 co-bound genomic regions with reduced accessibility following PAX8 depletion. Median ATAC-seq signal from 786-M1A cells expressing a control RNAi construct (shRen, N = 6) or PAX8-targeting RNAi constructs (shPAX8, N = 6). Median HIF2A and PAX8 ChIP-seq signals from 786-M1A and OS-LM1 xenografts, three tumous each. Asterisk indicates a region of interest. h, Fraction of PAX8 peaks (red) in all high-confidence open chromatin regions (all) and HIF2A ChIP-seq peaks in 786-M1A and OS-LM1 xenograft tumors. Asterisk indicates P < 1.0 × 10−300, two-sided Fisher’s exact test. Credit: Nature (2022). DOI: 10.1038/s41586-022-04809-8
An international group of researchers investigated why mutations often associated with renal cancer result specifically in renal cancer, instead of other cancer types.
Typically, gene mutations occurring in cancers are associated with only certain subtypes of cancer. The reasons for this have remained largely unclear.
According to the study published in Nature, the ability of mutations associated with renal cancer to drive the onset of cancer is based on proteins known as transcription factors, which regulate the normal functions of healthy renal cells.
"The study focuses on renal cancer, but the same principle may be applicable to cancers more generally. Cancer is often an incurable disease, and new therapies are needed. We identified factors whose inhibition slowed down the growth of renal cancer in experimental models. These molecules are potential drug targets," says Sakari Vanharanta, the project's principal investigator at the University of Helsinki.
A common gene mutation increases the risk of renal cancer
The mechanisms examined in the study concern clear cell renal cell carcinoma, the most common subtype of renal cancer. Most Europeans carry a genetic variant that increases the risk of developing this disease.
In the study, methods of experimental cancer biology were utilized in conjunction with targeted patient specimen analysis. The researchers tested whether transcription factors that control the tissue-specific functions of normal renal cells are also needed for the growth of renal cancer.
The results indicate that the ability of mutations associated with renal cancer to promote tumor formation was dependent on transcription factors that are particularly active in normal renal cells. When renal lineage-specific transcription factors were experimentally inactivated, cancer mutations were no longer able to activate genes relevant to tumor growth.
"The role of of genetic alterations, or mutations, in carcinogenesis is well established, but what we often don't know is how cancer mutations cause cancer. This makes it difficult to exploit the genetic information in cancer therapy. Understanding the functional consequences of cancer-related mutations is therefore important, and it can, as it has already in some cases, help scientists develop new cancer drugs," Vanharanta says.
More information: Saroor A. Patel et al, The renal lineage factor PAX8 controls oncogenic signalling in kidney cancer, Nature (2022). DOI: 10.1038/s41586-022-04809-8
Journal information: Nature
Provided by University of Helsinki