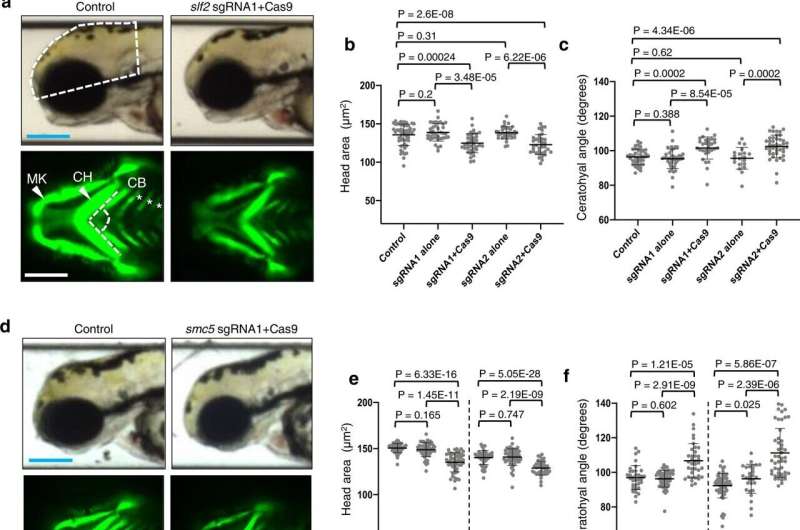
Loss of slf2 and smc5 in zebrafish give rise to microcephaly and aberrant craniofacial patterning. a Top: Representative lateral bright field images acquired at 3 days post-fertilization (dpf); white dashed shape depicts head size measured. Bottom: Representative ventral images of GFP signal from the anterior region of −1.4col1a1:egfp transgenic reporter larvae at 3 dpf. The white dashed lines show the ceratohyal angle. b Quantification of lateral head size measurements. Larvae were injected with two independent sgRNAs targeting slf2 with or without Cas9; n = 3 independent experiments (left to right; 56, 37, 37, 36, 36 larvae/batch). c Quantification of the ceratohyal angle. Larvae were injected with two independent slf2 sgRNAs: n = 3 independent experiments (left to right; 39, 42, 30, 20, 44 larvae/batch). d Top: Representative lateral bright field images at 3 dpf. Bottom: Representative ventral images of GFP signal in the anterior region of −1.4col1a1:egfp smc5 sgRNA1 transgenic larvae at 3 dpf. e Quantification of lateral head size measurements in 3 dpf larvae (as shown in panel a); n = 3 independent experiments (left to right; 50, 50, 52, 46, 53, 38 larvae/batch). The chart shows two independent experiments for sgRNA1 and sgRNA2 with a vertical line grouping independent controls with test conditions. f Quantification of the ceratohyal angle. Larvae were injected with two independent smc5 sgRNAs: n = 3 independent experiments (left to right; 34, 53, 37, 62, 28, 48 larvae/batch). The chart shows two independent experiments for sgRNA1 and sgRNA2 with a vertical line grouping independent controls with test conditions. g Left: Representative lateral bright field images of WT control and slf2−/− mutants at 3 dpf. Right: Quantification of lateral head size measurements in 3 dpf WT control and slf2−/− mutant larvae (as shown in a); n = 3 independent experiments (left to right; 10, 12, 12 larvae/batch). In (a, b): (top left) white dashed shape depicts head size measured; (bottom left) white dashed lines show the ceratohyal angle measured. MK Meckel’s cartilage, CH ceratohyal cartilage (indicated with arrowheads, respectively), and CB ceratobranchial arches (asterisks). Scale bars represent 300 μm, with equivalent sizing across panels. Error bars represent standard deviation of the mean. Statistical differences were determined with an unpaired Student’s t test (two sided). Credit: Nature Communications (2022). DOI: 10.1038/s41467-022-34349-8
Northwestern Medicine scientists have identified disease-causing DNA variants in two genes linked to neurodevelopmental deficits including microcephaly in children, according to findings published in Nature Communications.
Microcephaly, a condition in which a baby's head is smaller than expected, is one common manifestation of several different neurodevelopmental disorders caused by disruptions in DNA replication, repair or cell division.
In collaboration with a global team of scientists, investigators compared whole exome sequencing data from 11 affected individuals from around the world with microcephaly, short stature, cardiac abnormalities and anemia. They found that DNA variants in two genes, SLF2 and SMC5, compromise protein stability and reduce the ability of cells to repair damaged DNA.
Next, they validated those findings in a zebrafish model.
"We inactivated the SLF2 and SMC5 orthologs in the zebrafish genome," said Erica Davis, Ph.D., associate professor of Pediatrics and of Cell and Developmental Biology and co-senior author of the study.
"We measured the head size of the zebrafish and found that there was a significant reduction compared to normal siblings. We also evaluated the patterning of the zebrafish's face by measuring the cartilage structures and saw something similar to what was in this group of children."
The findings will aid in the early detection and diagnosis of microcephaly and other conditions, Davis said.
"The findings from this research will add two genes to diagnostic gene panel tests or clinical whole exome or genome sequencing tests that are being ordered for kids who present with microcephaly or a concern for neurodevelopmental disease," said Davis, who is also the Anne Marie and Francis Klocke, MD Research Scholar at Ann & Robert H. Lurie Children's Hospital of Chicago.
"It's also informing us about groups of proteins that perform a similar function in the cell, the RAD18-SLF1/2-SMC5/6 complex."
While the complex is not yet well understood, Davis said, the findings of the study underscore the importance of these genes in maintaining genome stability by regulating both DNA replication and cell division.
Further research will focus on better understanding this connection and why the brain is so susceptible, Davis said. "A major theme of my lab is an interest in ultra-rare congenital disorders," Davis said.
"The first pillar of that is identifying the causal genes, the second is understanding the mechanism and the third is therapeutic development. As we develop these zebrafish models, this gives us a new tool to try to identify ways to correct a cellular process that's gone wrong."
More information: Laura J. Grange et al, Pathogenic variants in SLF2 and SMC5 cause segmented chromosomes and mosaic variegated hyperploidy, Nature Communications (2022). DOI: 10.1038/s41467-022-34349-8
Journal information: Nature Communications
Provided by Northwestern University