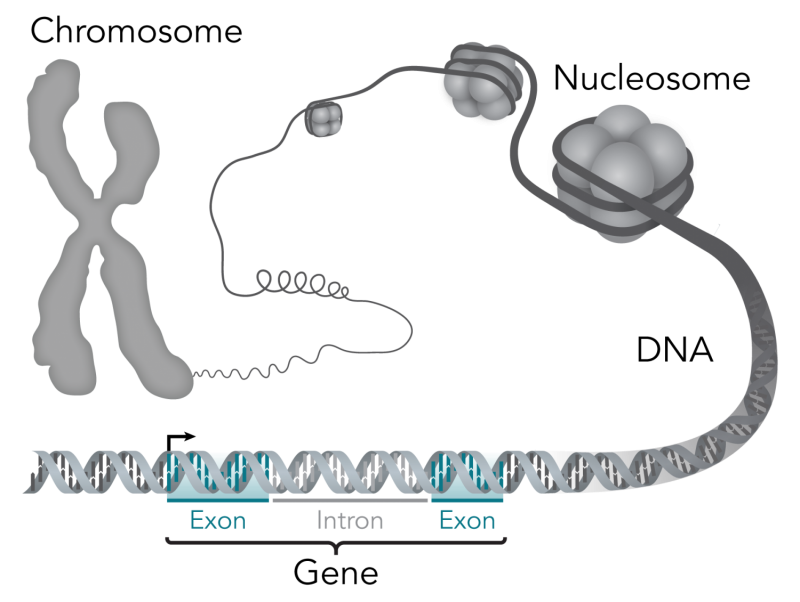
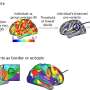
This stylistic diagram shows a gene in relation to the double helix structure of DNA and to a chromosome (right). The chromosome is X-shaped because it is dividing. Introns are regions often found in eukaryote genes that are removed in the splicing process (after the DNA is transcribed into RNA): Only the exons encode the protein. The diagram labels a region of only 55 or so bases as a gene. In reality, most genes are hundreds of times longer. Credit: Thomas Splettstoesser/Wikipedia/CC BY-SA 4.0
In a new paper published online by Nature Genetics, researchers from Dana-Farber Cancer Institute (DFCI) and the Broad Institute systematically investigated somatic copy number alterations of noncoding regions across cancers, integrating genomic, epigenomic, and transcriptomic data.
The team found six super-enhancer regions that are focally amplified across different cancer types, including two that are associated with overexpression of the MYC oncogene, suggesting that this type of modification may be a common mechanism activating cancer driver genes.
The team, which was led by senior author Matthew Meyerson and first authors Xiaoyang Zhang, Peter Choi, and Joshua Francis – all of Broad and DFCI – also used genome-editing technologies to validate the oncogenic function of these focally amplified super-enhancers.
More information: Xiaoyang Zhang et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers, Nature Genetics (2015). DOI: 10.1038/ng.3470
Journal information: Nature Genetics
Provided by Broad Institute of MIT and Harvard