Credit: NIAID via Flickr
A research team led by scientists from the Broad Institute has uncovered a novel group of chemical inhibitors that can kill the bacteria that causes tuberculosis (TB). Importantly, these chemical compounds take aim at a previously unexplored molecular target, paving a potential path toward an entirely new class of antibiotics, which are profoundly needed to combat the worldwide problem of drug resistance in TB.
The researchers' new chemical finds, which are described in Nature, flow from an innovative, high-throughput drug discovery approach dubbed PROSPECT that can unearth promising drug candidates often missed by conventional methods. Importantly, this new strategy also provides key early insights into how the potential drugs work—for example, do they aim at molecular targets in ways that mimic existing anti-TB drugs, or do they represent fresh new forays into the microbe's biochemical universe?
"The rising tide of antibiotic resistance is an urgent global health threat, particularly for tuberculosis," said senior author Deborah Hung, a core institute member of the Broad Institute and co-director of its Infectious Disease and Microbiome Program. "With PROSPECT, we've identified scores of chemical compounds with potency against TB that cover a diverse range of molecular targets—many of which would not have been found if we had used the standard discovery methods."
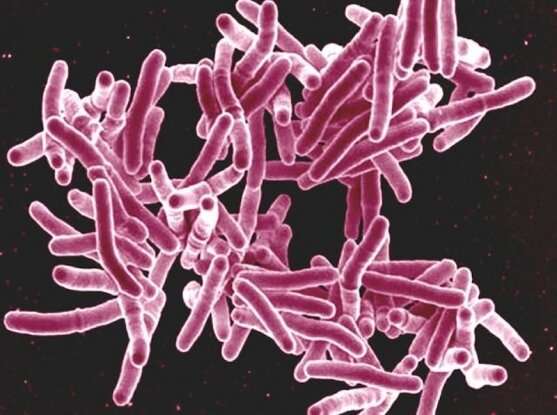
According to the World Health Organization, antibiotic resistance ranks among the top health threats across the globe. For TB, the problem is particularly severe. As the leading infectious disease killer of adults in the world, the bacteria that causes TB, Mycobacterium tuberculosis, has a frightening reach: In 2017, roughly 10 million people developed an active infection and some 1.6 million people died. Current TB treatments must be taken over the course of several months and sometimes fail to fully eradicate the culprit bacteria. Those shortcomings have spurred the emergence of drug-resistant forms of the disease, which are now spreading in many parts of the world.
"In order to counteract this worrying trend, we need to ensure the clinical pipeline for anti-TB drugs is sufficiently full and includes drug candidates that work in ways that are distinct from the old drugs," said Hung, who is also an infectious disease physician at Brigham and Women's Hospital and Massachusetts General Hospital.
To address these needs, Hung and her colleagues, including first author Eachan Johnson, developed PROSPECT (PRimary screening Of Strains to Prioritize Expanded Chemistry and Targets), an approach that relies on a unique collection of TB strains that have been intentionally sensitized to drug treatment by molecularly crippling (but not fully disabling) each of the hundreds of essential genes in M. tuberculosis (genes without which the bacterium cannot survive). These so-called hypomorphic strains are each marked with a unique molecular barcode so they can be pooled and analyzed together, and then later distinguished from one another. Engineering the strains, numbering over 450 in total, was a major effort involving collaborators at several organizations: Sabine Ehrt and Dirk Schnappinger at Weill Cornell Medical College, Eric Rubin at the Harvard T. H. Chan School of Public Health, and Christopher Sassetti at the University of Massachusetts Medical School.
Johnson and his colleagues then used these pooled strains as the basis for a large-scale drug screening effort, which included a total of 150 hypomorphic strains and more than 50,000 chemical compounds, searching for chemicals that killed the weakened bacteria but left normal bacteria unscathed.
After systematically mining the resulting data—essentially, millions of pairwise interactions between different chemicals and essential genes—the researchers uncovered hundreds of compounds with previously unknown anti-TB activity. In the Nature paper, they focus their initial analyses on a subset that includes 45 compounds. For the most promising ones, they worked to optimize the chemicals' molecular structures in order to render the compounds lethal even for normal, non-vulnerable bacteria.
"The real beauty of PROSPECT is that it enables the fairly rapid discovery of 10 times more compounds with potency against TB—candidate drugs that might otherwise be missed by conventional drug screening methods," said Johnson. "It also allows us to determine the compounds' mechanism of action so we can prioritize them based not just on their ability to kill bacteria but also on how they operate molecularly."
The compounds that emerged from this effort cover a range of molecular targets, including several that have been previously identified. These include enzymes involved in basic cellular processes such as DNA replication and transcription, as well as other targets involved in the generation of critical nutrients as well as the bacteria's tough outer shell.
But perhaps the most surprising finds were a set of compounds that target the EfpA gene, which is thought to function as a kind of molecular pump that expels substances from within the bacteria into their environment. No drugs or drug candidates have been previously discovered that neutralize this pump, underscoring the novelty of the team's approach.
Johnson, Hung, and their colleagues are now working to further optimize these novel compounds targeting EfpA, chemically modifying them so they have more drug-like properties, with the goal of advancing to early-stage pre-clinical testing in animal models and, eventually, human clinical trials.
More information: Tthe Broad Institute's Infectious Disease and Microbiome Program: www.broadinstitute.org/node/8543/
Johnson EO, et al. Large-scale chemical-genetics yields new M. tuberculosis inhibitor classes. Nature. DOI: 10.1038/s41586-019-1315-z
Journal information: Nature
Provided by Broad Institute of MIT and Harvard