Every tumour is unique and requires specific treatment. A thorough and complete analysis of the genetic activity in the tumour cells is necessary to determine the appropriate treatment. Researchers at TU Delft, in collaboration with researchers from Columbia University and the Antoni van Leeuwenhoek Hospital have achieved significant improvements in this type of analysis. The results were published on 4 and 10 April in the scientific journals PNAS and PLOS Genetics.
One way of identifying new cancer genes is to use viruses. They can insert their genetic material into the DNA of an organism, for example in mice. These changes in the genetic material – mutations – cause certain tumours to develop in the mice. By carrying out a genetic analysis of the tumours and identifying the mutations, it is possible to discover which genes caused a tumour.
Gene expression analysis
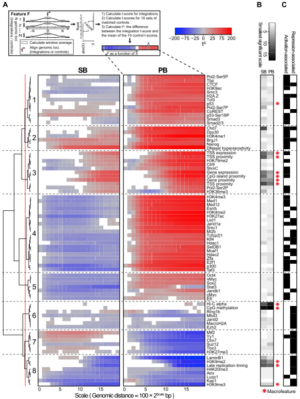
'We combined this technique with gene expression analysis', explains Dr Jeroen de Ridder, a bioinformatics specialist at the Delft Bioinformatics Lab of TU Delft. 'The gene expression profile shows the activity of all genes in the tumour. We can see whether a certain gene is switched 'on' or 'off', or in between. This means we can characterise precisely how mutations affect the activity of the genes in the tumour.'
De Ridder adds: 'This research principle is not new, but the added value of the recent advances is the completeness of the analysis.' At Delft, our work has focussed on the informatics side of the research. Using a regression model we can now identify the effect of a mutation on all genes at the same time. This has provided many new insights into the effect of mutations on gene activity. In addition, by means of statistical analyses and the use of existing data sets, greater insight has been gained into which parts of the DNA are susceptible to mutations of this type. On that basis we reached the conclusion that current analysis methods relatively often produce incorrect results.
'In the medical world there is an increasing awareness that each tumour is unique and therefore in fact requires specific treatment. Our method increases the possibilities and the understanding required to develop individual treatment for each tumour.'
More information: "Identifying regulatory mechanisms underlying tumorigenesis using locus expression signature analysis." Eunjee Lee, Jeroen de Ridder, Jaap Kool, Lodewyk F. A. Wessels, Harmen J. Bussemaker . PNAS 2014 ; published ahead of print April 2, 2014. www.pnas.org/content/early/201 … 293111.full.pdf+html
"Chromatin Landscapes of Retroviral and Transposon Integration Profiles." Johann de Jong, Waseem Akhtar, Jitendra Badhai, Alistair G. Rust, Roland Rad, John Hilkens, Anton Berns, Maarten van Lohuizen, Lodewyk F. A. Wessels, Jeroen de Ridder. PLOS Genetics. Published: April 10, 2014.
Journal information: PLoS Genetics , Proceedings of the National Academy of Sciences
Provided by Delft University of Technology