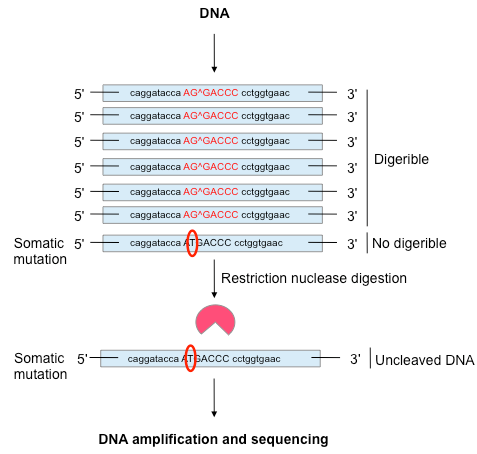
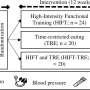
DNA amplification and sequencing. Credit: Centre for Molecular Biology
It has been proposed that somatic gene variations (SNV) present in few brain cells could facilitate the development of neurodegenerative disorders like Alzheimer's disease. Testing that hypothesis requires DNA sequencing directly in brain cells or tissue rather than in blood cells.
However, the identification of SNV by standard and reliable sequencing procedures does not work well when the number of cells bearing the specific SNV (or mutation) is very low within the tissue. In this way, another techniques, such as high-throughput methods, could be used. However, those methods can introduce errors in reading sequence alignments that can interfere with the identification of true somatic variations.
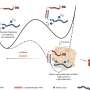
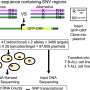
In this work, once a new SNV was identified using high-throughput methodologies, the bulk DNA lacking that particular SNV was eliminated by the use of a restriction nuclease that cleaves bulk DNA molecules but not the ones containing the specific SNV. In further steps, the uncleaved DNA was amplified and sequenced by a reliable (error-free) technique such as the Sanger's sequencing method (see Figure). By following this approach we have been able to achieve the validation of some somatic brain mutations.
More information: Alberto Gomez-Ramos et al, Validation of Suspected Somatic Single Nucleotide Variations in the Brain of Alzheimer's Disease Patients, Journal of Alzheimer's Disease (2017). DOI: 10.3233/JAD-161053
Journal information: Journal of Alzheimer's Disease
Provided by IOS Press