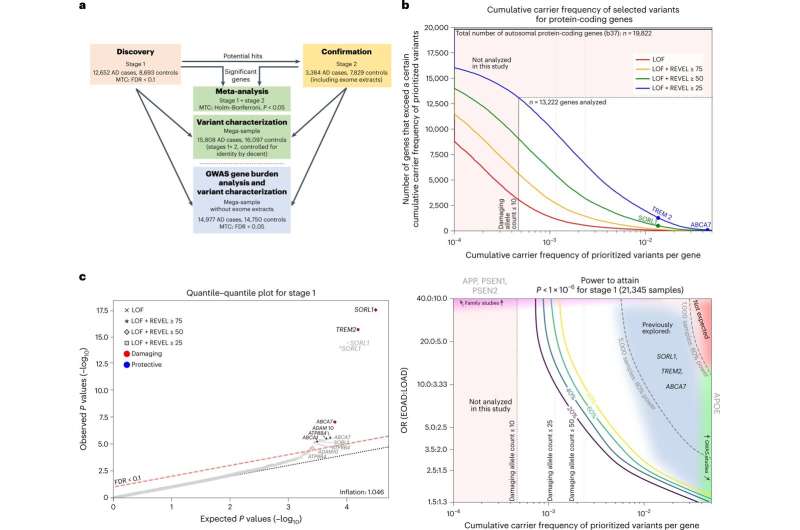
Study setup and power. a, Schematic of the study setup. The AD association of genes identified in stage 1 was confirmed in stage 2 and significance was determined by meta-analysis. Variant characteristics were investigated in a merged mega-sample rather than the meta-sample, allowing more accurate variant effect size estimates for variant categories/age-at-onset bins. The mega-sample (without exome extracts) was also used for the GWAS gene burden analysis. MTC, multiple testing correction. b, Top, number of genes (y axis) with at least a certain cumulative carrier frequency of prioritized variants (x axis), prioritized according to different deleteriousness thresholds. White box, genes with a cMAC ≥ 10 (cumulative minor allele count of ≥10 prioritized alleles identified across the 12,652 cases and 8,693 controls in the stage 1 sample) were considered to have sufficient carrier frequency to allow burden analysis. The SORL1, TREM2 and ABCA7 genes are indicated, revealing that carriers of rare damaging variants in these genes are relatively common, allowing identification in smaller sample sizes . Bottom, power analysis for stage 1, to attain a P < 1 × 10−6, at the same scale as the top figure. For comparison, we indicate 80% power thresholds for sample sizes of 1,000 and 5,000 individuals (subsampled from stage 1). Cumulative carrier frequency and estimated effect size ranges are indicated for common variants identified to associate with AD by GWAS (green), rare-variant burdens in SORL1, TREM2 and ABCA7 identified using sequencing studies (gray/blue), and for rare variants observed in autosomal dominant AD (magenta). Common variants with high effect sizes (red) are not expected to exist. Genes with cMAC < 10 were not analyzed (pink). Power calculations show that aggregating more cases and controls might allow for the identification of rare-variants that have a large effect on AD but for which only few carriers are observed, or for variants that have a modest/average effect on AD, for which many carriers are observed c, Quantile–quantile plot of P values determined in the stage 1 discovery analysis based on an ordinal logistic burden test. For each of 13,222 genes, we tested the burden of variants adhering to four variant deleteriousness thresholds, conditional on having a cMAC ≥ 10 (n = 31,204 tests). Threshold for multiple testing correction: FDR < 0.1, P value inflation, 1.046. Gene names in black indicate the deleteriousness threshold of the most significant burden test in that gene. Credit: Nature Genetics (2022). DOI: 10.1038/s41588-022-01208-7
Two new genes that raise a person's risk of developing Alzheimer's Disease have been discovered by researchers.
An international team, involving Cardiff University's Dementia Research Institute, compared 32,000 genetic codes from patients with Alzheimer's disease and healthy individuals.
The research uncovered several new genes and specific mutations in those genes that lead to the development of Alzheimer's disease. They found rare, damaging genetic mutations in the genes known as ATP8B4 and ABCA1 which could lead to an increased risk of Alzheimer's disease.
The researchers also found evidence of genetic alternation in a further gene, ADAM10.
Professor Julie Williams, Director of the Dementia Research Institute at Cardiff University, and a co-author on the study said, "These findings point us towards very specific processing in the brain, which includes differences in the brain's immune system and how the brain processes cholesterol. These differences impact brain functioning and leads to the development of Alzheimer's disease."
Alzheimer's disease is the most common form of dementia in the UK. An estimated 60–80% of the risk of Alzheimer's disease can be explained by genetic factors. For early onset Alzheimer's (under 65 years), this increases to more than 90%.
Professor Williams added, "This study helps expand our knowledge about who is at risk of developing this form of dementia. These genetic discoveries also allow us to understand the mechanisms underlying Alzheimer's, as well as create genetic models of the disease to develop targeted therapies in the future—through new drug-based treatments or even gene therapy.
"The Dementia Research Institute at Cardiff University is well-placed to apply this research and take it forward into developing disease models. We are the biggest investment in dementia research in Wales, with over 100 researchers focused on developing our understanding of dementia and delivering new treatments.
The study, "Exome sequencing identifies rare damaging variants in ATP8B4 and ABCA1 as risk factors for Alzheimer's disease," is published in Nature Genetics.
More information: Henne Holstege et al, Exome sequencing identifies rare damaging variants in ATP8B4 and ABCA1 as risk factors for Alzheimer's disease, Nature Genetics (2022). DOI: 10.1038/s41588-022-01208-7
Journal information: Nature Genetics
Provided by Cardiff University