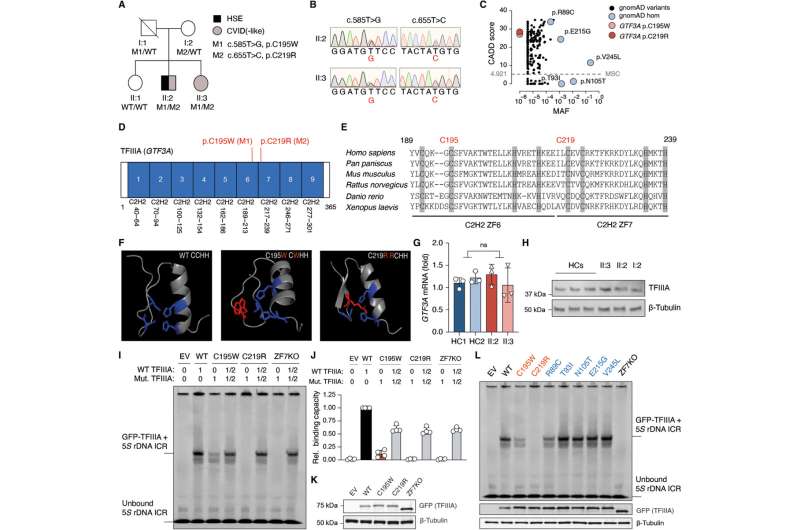
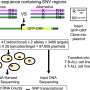
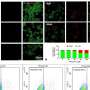
Mutations in GTF3A result in impaired DNA binding ability in a patient with CVID and HSE. (A) Family pedigree with clinical phenotype and allele segregation of GTF3A variants. (B) Electropherograms of genomic DNA of primary fibroblasts from II:2 and II:3. (C) CADD score against minor allele frequency (MAF) for GTF3A variants in gnomAD with MSC (99% confidence interval) of 4.921. (D) Schematic of TFIIIA domain structure comprising nine C2H2 ZFs, with p.C195W and p.C219R mutations shown in red. Numbers below indicate amino acids. (E) Clustal Omega sequence alignment of TFIIIA (ZF6 and ZF7) from the indicated species, with conserved C2H2 residues shown in gray. Numbers above indicate amino acids. (F) Predicted 3D structure of the C2H2 ZFs of human TFIIIA protein (WT and mutants). Missense mutations (p.C195W and p.C219R) are shown in red. (G) RT-qPCR analysis of GTF3A expression in fibroblasts from II:2 and II:3 compared with HCs. (H) Endogenous TFIIIA protein expression in fibroblasts from HCs, II:2, II:3, and I:2, assessed by IB. (I) TFIIIA binding to 5S rDNA ICR assessed by EMSA on WCLs of HEK293T cells that were transfected for 16 hours with empty vector (EV) or GFP-fused TFIIIA WT, missense mutants (C195W and C219R), or ZF7KO (control) at indicated ratios. (J) Densitometric quantification of (I). (K) Expression of WT and mutant TFIIIA in the WCLs for (I), determined by IB. (L) TFIIIA binding to 5S rDNA ICR, determined by EMSA on WCLs of HEK293T cells that were transfected for 16 hours with EV or the indicated GFP-fused TFIIIA WT and mutant constructs. Each data point represents one biological replicate from three (G) or four (J) independent experiments (mean ± SD), or data shown are representative of at least two independent experiments (H, I, K, and L) (two-tailed, unpaired Student’s t test). ns, statistically not significant. Credit: Science Immunology (2022). DOI: 10.1126/sciimmunol.abq4531
A collaboration between Ghent University in Belgium and Cleveland Clinic's Florida Research & Innovation Center (FRIC) found a new way genetics influences the body's antiviral response by studying a life-threatening disease caused by a common virus: herpes simplex virus 1 (HSV-1).
The researchers analyzed genetic data from a patient with immunodeficiency and hospitalized at nine months old with herpes encephalitis, a rare but life-threatening brain inflammation after HSV-1 infection. They identified novel mutations in the gene GTF3A, and found that these mutations impair the innate immune response.
The findings, published in Science Immunology, hold potential as a genetic marker doctors could use to gauge a child's risk of herpes encephalitis, although such mutations are generally very rare in the population.
Many people are infected in childhood with the HSV-1 virus but the vast majority don't suffer from encephalitis. The most common symptom of HSV-1 is oral cold sores, but many people show no signs at all. HSV-1 is more threatening to children and adults who are immunodeficient, whose immune system cannot control the virus well.
"Genetic and mechanistic analyses of uncommon viral diseases like herpes encephalitis are quite rare. In fact, the causes underlying severe herpes encephalitis are often unknown," says Michaela Gack, Ph.D., FRIC's scientific director. "This information provides us with invaluable insight into the fundamental molecular processes that govern our immune response and opens up opportunities for future research on severe disease outcomes."
The Ghent research team led by Filomeen Haerynck, M.D., Ph.D., reached out to Dr. Gack's team after finding the mutations in the gene. Dr. Gack's lab studies interactions between the human immune system and viruses on a molecular level.
The GTF3A mutations shape how cells respond to viral activity through the genetic makeup of a protein called TFIIIA. TFIIIA plays a role in helping a human enzyme produce certain types of RNA that can determine specific functions inside cells. Some RNAs can elicit an anti-herpes viral immune response.
Dr. Gack's team tested cells that have the mutations, and found that because of defects in certain immunostimulatory RNAs, the cells were more susceptible to HSV-1 infection and lost the ability to control the HSV-1 virus.
The affected gene is part of the body's defense system that produces interferons to combat viruses. Interferons are crucial to the human immune response and for suppressing virus infection and spread.
This new genetic pathway could be helpful in understanding the immune response to other viruses, like Epstein-Barr virus, a common virus linked to mononucleosis and associated with certain types of cancer and multiple sclerosis.
"Understanding the molecular processes underlying antiviral responses is key to treating or possibly preventing severe viral infections that change patients' and families' lives," Dr. Gack said. "Our findings on critical immune defense proteins may translate into new therapies in the future."
More information: Leslie Naesens et al, GTF3A mutations predispose to herpes simplex encephalitis by disrupting biogenesis of the host-derived RIG-I ligand RNA5SP141, Science Immunology (2022). DOI: 10.1126/sciimmunol.abq4531
Journal information: Science Immunology
Provided by Cleveland Clinic