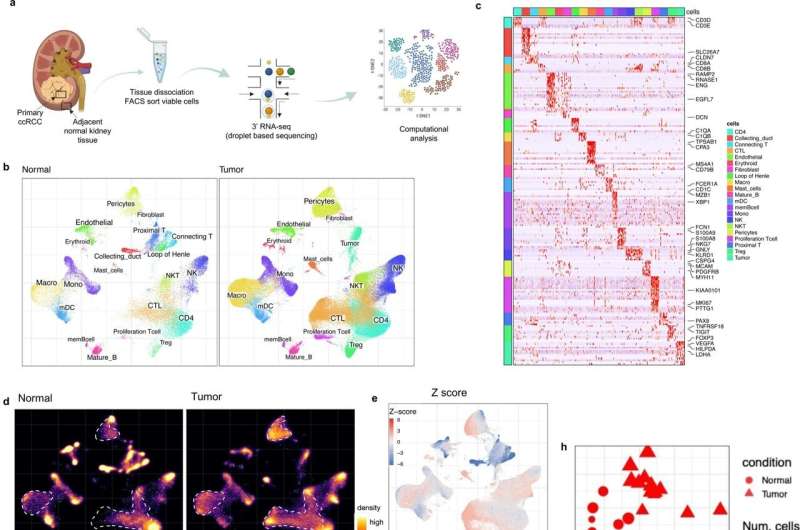
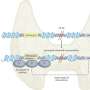
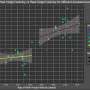
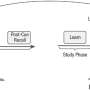
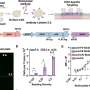
Single-cell landscape of the ecosystem in primary ccRCC and adjacent normal tissue. a Experimental design created with Biorender.com and Adobe Illustrator. b Integrative analysis of scRNA-seq samples from 26 RCC samples, visualized using a common UMAP embedding for adj-normal (left) and tumor kidney tissue (right). c Heatmap showing expression of markers for major cell populations. d Changes in the composition of all compartments combining all sample fractions and is visualized as cell density on the joint embedding. e Statistical assessment of the cell density differences comparing tumor with adjacent normal. A two-side Wilcoxon test was used, visualized as a Z score. Red indicates increased cell abundance in tumor, blue indicates decreased cell abundance in tumor. f Change in cell composition evaluated by Compositional Data Analysis. The x-axis indicates the separating coefficient for each cell type, with the positive values corresponding to increased abundance in tumor, and negative to decreased abundance. The boxplots and individual data points show uncertainty based on bootstrap resampling of samples and cells (see Methods). Boxplot includes center line: median; box limits: upper and lower quartiles; whiskers extend at most 1.5× interquartile range past upper and lower quartiles. g The boxplots showing the magnitude of transcriptional change between primary RCC and normal kidney tissue in major cell populations. The magnitude is assessed based on a Pearson linear correlation coefficient, normalized by the medium variation within primary RCC and normal kidney tissue (see Methods). Statistics significance within each cell type is measured with permutation test in sample group (Pericytes **p = 0.003; Endothelial **p = 0.003; Fibroblast **p = 0.003; Erythroid **p = 0.005; Macro **p = 0.004; Proliferation T cell *p = 0.01; Treg **p = 0.009). Boxplot includes center line: median; box limits: upper and lower quartiles; whiskers extend at most 1.5× interquartile range past upper and lower quartiles. h MDS embedding of different samples, based on their overall expression distance. The similarity measure measures the magnitude of expression change for each subpopulation, using size-weighted average to combine them into an overall expression distance that controls the compositional differences. Shape indicates different sample fractions. Credit: Nature Communications (2022). DOI: 10.1038/s41467-022-33375-w
Among patients with kidney cancer, the activity of four specific genes in the cancer cells seems to be able to predict the risk of the tumor spreading and the patient's chances of survival. This is shown by researchers from Karolinska Institutet in Sweden in a preclinical study published in Nature Communications.
"This could potentially become a tool to gain a better understanding of the course of the disease at an early stage. Patients with a cancer profile with a high probability of spreading could then be monitored more closely, to quickly detect and treat any growth of the tumor," says Ninib Baryawno, senior researcher at the Department of Women's and Children's Health, Karolinska Institutet, and the study's last author.
Clear cell kidney cancer is the most common form of kidney cancer in adults. If the tumor is confined to the kidneys, the prognosis is often favorable, but if it has spread to the skeleton, which occurs in about a third of patients, the five-year survival rate is only about 10%.
Immunotherapy known as checkpoint inhibitors have in recent years become an important treatment for patients with clear cell kidney cancer. But it is common for the cancer cells to develop resistance to the treatment, which may partly be attributed to factors in the environment around the cancer cells, the so-called tumor microenvironment.
In the current study, the researchers examined samples from nine patients with clear cell kidney cancer. The study is a collaboration between researchers at Karolinska Institutet, clinicians at Massachusetts General Hospital, where the patients were recruited, and computational scientists from Harvard Medical School in Boston, USA.
Both tumor tissue and nearby normal kidney tissue were collected from the same patient to be able to make matched comparisons and control for inter-individual variation. The cells were studied by single-cell analysis; a sequencing technique that makes it possible to investigate each single cell in the tissue and the gene expression, that is, which genes are active, in individual cells.
In two patients, the researchers also compared primary tumor tissue from the kidney with tissue from skeletal metastases.
The study demonstrates that a genetic signature consisting of four specific genes is predictive of whether the tumor will spread to the skeleton and of survival. Simultaneous overexpression of these genes (SAA1, SAA2, APOL1 and MET) suggests that the patient has a greater risk of developing a tumor that spreads and a poorer survival outcome.
The association of the gene signature to the risk of spreading was also confirmed when the researchers examined tumor cells from bone metastases in seven patients with metastatic clear cell kidney cancer.
Moreover, the study shows that the microenvironment of the tumor inhibits the immune system, and the researchers suggest several possible targets for drugs that may be interesting to investigate further. These were identified with computer simulations of cell interactions.
The study provides important biological knowledge about the interaction between tumor cells and their microenvironment in clear cell kidney cancer, the researchers say.
"We hope that our results will contribute to further investigations of factors that affect the tumor microenvironment, which can ultimately provide new ways to treat relapse and the spread of cancer. For us, the next step is to study how metastases in the bone marrow and the skeleton differ from the local tumor in the kidney but also how the bone marrow in patients with kidney cancer metastases in the skeleton differs from healthy bone marrow. We hope that it can help us answer the question as to why immunotherapy does not work in some kidney cancer patients," says Adele Alchahin, Ph.D. student at the Department of Women's and Children's Health, Karolinska Institutet, and one of the study's first authors.
The KI researchers involved in the publication state that there are no potential conflicts of interest. Other authors have links to various pharmaceutical and biotechnology companies in the form of, among other things, founding and consulting commitments. See the scientific article for further information.
More information: Adele M. Alchahin et al, A transcriptional metastatic signature predicts survival in clear cell renal cell carcinoma, Nature Communications (2022). DOI: 10.1038/s41467-022-33375-w
Journal information: Nature Communications
Provided by Karolinska Institutet