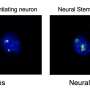
Protein changes at brain cell synapses from patients with schizophrenia were strikingly similar to those from patients with bipolar disorder. Credit: Sonja Vasiljeva/Broad Institute
Although bipolar disorder and schizophrenia are diagnosed as distinct psychiatric conditions, both are considerably heritable with molecular roots that are poorly understood. Some people diagnosed with one disorder have symptoms and clinical features in common with the other, supporting the notion that the conditions lie on a spectrum. And human genetics studies have suggested that the junctions between brain cells, known as synapses, play a key role in both conditions.
New research has taken a deep look at the role of brain cell synapses in the two conditions and has found molecular evidence suggesting that the disorders may not be as different as once thought. Scientists in the Stanley Center for Psychiatric Research at the Broad Institute of MIT and Harvard collaborated with experts in the Broad's Proteomics Platform to measure thousands of proteins present at synapses in the brains of people with schizophrenia, those with bipolar disorder, and those unaffected by either illness.
Their analysis revealed changes in synapse proteins that were remarkably similar in the two conditions. In mice with a mutated gene that's been linked to both conditions, the scientists found that related biochemical pathways were similarly altered. Described in Cell Reports, the work sheds new light on what goes awry at a molecular level in these conditions and presents a data resource for further exploration of their shared roots.
"Many lines of evidence point to the role of the synapse in these conditions," said study co-senior author Borislav Dejanovic, a scientist in the Stanley Center who is now director at Vigil Neuroscience. "With refined experimental methods and proteomics capabilities, we're fortunate to be able to examine this connection at an unprecedented scale and resolution."
"Our findings support the Stanley Center's goal of better understanding the pathogenic mechanisms underlying bipolar disorder and schizophrenia," said co-senior author and Broad core institute member Morgan Sheng, who is also co-director of the Stanley Center, a professor of neuroscience in MIT's Department of Brain and Cognitive Sciences, and an affiliate of the Picower Institute for Learning and Memory. "The next steps are to examine these pathways in normal brain function and to explore how they might go wrong in psychiatric conditions."
Synaptic surprises
Isolating and purifying synapses from brain tissue samples is no easy task, but that is what the team had to do to analyze the structure's proteins—its working parts—at a large scale. They carried out a labor-intensive protocol in the lab that Sheng and Dejanovic have helped to develop and refine over the past many years.
In the current study, the researchers purified synapses from postmortem tissue from an area of the brain known as the dorsolateral prefrontal cortex, from 35 individuals who had been diagnosed with schizophrenia, 35 people with bipolar disorder, and 35 unaffected individuals. Collaborators in the Proteomics Platform analyzed the purified synapses by mass spectrometry, measuring the abundance of thousands of proteins and protein fragments in an unbiased way.
"We're able to make these discoveries because we can get such deep, sensitive, and quantitative coverage of the synaptic proteome, thanks to our team members in the Broad's Proteomics Platform," said Dejanovic.
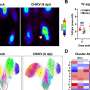
In samples from people with each disorder, the team observed changes in the levels of hundreds of proteins compared to control individuals. Strikingly, more than 200 of these proteins were either enriched or depleted similarly in both conditions. The scientists were surprised by how comparable the patterns of protein changes were between the disorders.
The team next examined whether the altered proteins were connected in known networks or biochemical pathways within the cell, through computational analyses led by study first author Sameer Aryal.
"Analyzing protein networks helps us understand how clusters of proteins change in disease synapses, revealing molecular connections that aren't easily discernible by looking only at individual protein levels," said Aryal, a postdoctoral researcher in the Sheng lab.
Some of the upregulated proteins were associated with autophagy (the cell's protein recycling process) and pathways that traffic molecules from one part of the cell to another. Downregulated proteins were related to synaptic, mitochondrial, and ribosomal function, suggesting that energy metabolism and protein production might be curbed in these cells.
Model mice
The researchers also observed similar pathway changes in mice deficient in Akap11, a gene that in humans is a risk factor for both schizophrenia and bipolar disorder, a discovery made by scientists at the Stanley Center and elsewhere in 2022. The pathway similarities in the mouse model reinforced the results in the human samples and validated the model as a useful experimental system for studying these conditions. "These are systems-wide brain disorders, so we need valid animal models like the Akap11 mutant mice that have a fully intact nervous system," Sheng explained.
Stanley Center scientists are currently examining the Akap11-deficient mice to elucidate the impact of these protein alterations, including changes in molecular trafficking at the synapse. The results of this unbiased analysis will help them and other research groups prioritize proteins and pathways for further study.
This study examined proteins from multiple synapses aggregated together, and synapses from only one brain region. The scientists said future work to study proteins from individual synapses could provide an even higher-resolution view. And analyzing synapses from other regions of the brain could reveal region-specific molecular alterations, which could help build a better understanding of these systems-wide disorders.
More information: Sameer Aryal et al, Deep proteomics identifies shared molecular pathway alterations in synapses of patients with schizophrenia and bipolar disorder and mouse model, Cell Reports (2023). DOI: 10.1016/j.celrep.2023.112497
Journal information: Cell Reports
Provided by Broad Institute of MIT and Harvard