Biomarkers for inflammatory disease
Gene-expression profiles might be used to identify prognostic biomarkers for Kawasaki disease, and help to unravel the underlying biology of the illness, research published this week in the online open access journal Genome Biology reveals. The new findings also support the idea that gene-expression profiles might be used to generate biomarkers for other systemic inflammatory illnesses.
Kawasaki disease, an acute, self-limited vasculitis, is the leading cause of acquired heart disease in children in developed countries, but its aetiologic and pathogenic mechanisms remain unclear.
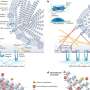
A team of researchers led by David Relman, Stanford University, US, and Jane Burns, University of California at San Diego, US, characterized the gene expression patterns that occur in the blood cells of patients with this disease. They examined genome-wide transcript expression patterns in the blood of 77 children with Kawasaki disease. The acute phase of the illness was accompanied by an increase in gene transcripts associated with innate immune mechanisms and proinflammatory responses, and a decrease in transcripts associated with natural killer cells and CD8+ lymphocytes, which help clear infected or abnormal cells from the body.
They showed that the transcript patterns during the acute phase of the disease varied dramatically with day of illness, and that differences in expression patterns between patients were associated with clinical parameters that physicians have used to manage and make predictions about the course of the disease. Patients who showed higher expression levels of specific transcripts (e.g., carcinoembryonic antigen-related cell adhesion molecule 1; CEACAM1) were less likely to respond to intravenous immunoglobulin, a highly effective but poorly understood treatment for preventing coronary artery aneurysms and reducing fever in Kawasaki disease.
This work contributes to our understanding of how the disease develops, how the treatment works, and how doctors might identify patients who are candidates for other therapies.
Source: BioMed Central