Discovery opens door for drugs to fight bird flu, other influenza epidemics
Researchers at Rutgers University and The University of Texas at Austin have reported a discovery that could help scientists develop drugs to fight the much-feared bird flu and other virulent strains of influenza.
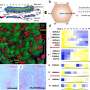
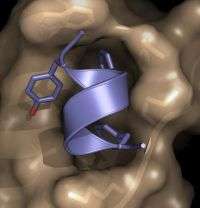
The researchers have determined the three-dimensional structure of a site on an influenza A virus protein that binds to one of its human protein targets, thereby suppressing a person's natural defenses to the infection and paving the way for the virus to replicate efficiently. This so-called NS1 virus protein is shared by all influenza A viruses isolated from humans – including avian influenza, or bird flu, and the 1918 pandemic influenza virus.
A paper detailing this breakthrough discovery appears in the PNAS (Proceedings of the National Academy of Sciences) Early Edition and will be published in an upcoming issue of the PNAS print edition.
About 10 years ago, Professor Robert M. Krug at The University of Texas at Austin discovered that the NS1 protein binds a human protein known as CPSF30, which is important for protecting human cells from flu infection. Once bound to NS1, the human protein can no longer generate molecules needed to suppress flu virus replication. Now, researchers led by Rutgers Professor Gaetano T. Montelione and Krug identified the novel NS1 binding pocket that grasps the human CPSF30 protein.
"Our work uncovers an Achilles heel of influenza A viruses that cause human epidemics and high mortality pandemics," said Montelione, professor of molecular biology and biochemistry. "We have identified the structure of a key target site for drugs that could be developed to effectively combat this disease."
X-ray crystallography, which was carried out by Kalyan Das, Eddy Arnold, LiChung Ma and Montelione, identified the three-dimensional structure of the NS1 binding pocket. "The X-ray crystal structure gives us unique insights into how the NS1 and human protein bind at the atomic level, and how that suppresses a crucial antiviral response," said Das, research professor at Rutgers.
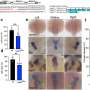
Rei-Lin Kuo, Jesper Marklund, Karen Twu and Krug at The University of Texas at Austin verified the key role of this binding pocket in flu replication by genetically engineering a change to a single amino acid in the NS1 protein's binding pocket, which in turn eliminated the protein's ability to grasp the human protein that is needed to generate antiviral molecules. These investigators then produced a flu virus with an NS1 pocket mutation and showed that this mutated virus does not block host defenses, and as a consequence has a greatly reduced ability to infect human cells.
"These experiments validate the NS1 pocket as a target for antiviral drug discovery," said Krug, professor and chair of molecular genetics and microbiology. "Because this NS1 pocket is highly conserved in all influenza A viruses isolated from humans, a drug targeted to the pocket would be effective against all human influenza A strains, including the bird flu."
This project was supported by two different institutes at the National Institutes of Health (NIH), demonstrating how several NIH initiatives can complement each other. Support for the Rutgers research was provided in part by the Protein Structure Initiative (PSI) of the NIH Institute of General Medical Sciences, a follow-on to the human genome project, which is providing large numbers of protein samples and three-dimensional structures of biologically important proteins to the broad scientific community.
"This work underscores the value of scientific collaborations between large-scale structural centers and individual biomedical research labs," said John Norvell, director of the PSI.
Source: University of Texas at Austin