Computational method predicts new uses for existing medicines
For the first time ever, scientists are using computers and genomic information to predict new uses for existing medicines.
A National Institutes of Health-funded computational study analyzed genomic and drug data to predict new uses for medicines that are already on the market. A team led by Atul J. Butte, M.D., Ph.D., of Stanford University, Palo Alto, Calif., reports its results in two articles in the Aug. 17 online issue of Science Translational Medicine.
"Bringing a new drug to market typically takes about $1 billion, and many years of research and development," said Rochelle M. Long, Ph.D., who directs the NIH Pharmacogenomics Research Network. "If we can find ways to repurpose drugs that are already approved, we could improve treatments and save both time and money."
The scientists drew their data from the NIH National Center for Biotechnology Information Gene Expression Omnibus, a publicly available database that contains the results of thousands of genomic studies on a wide range of topics, submitted by researchers across the globe. The resource catalogs changes in gene activity under various conditions, such as in diseased tissues or in response to medications.
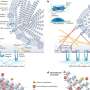
Butte's group focused on 100 diseases and 164 drugs. They created a computer program to search through the thousands of possible drug-disease combinations to find drugs and diseases whose gene expression patterns essentially cancelled each other out. For example, if a disease increased the activity of certain genes, the program tried to match it with one or more drugs that decreased the activity of those genes.
Many of the drug-disease matches were known, and are already in clinical use, supporting the validity of the approach. For example, the analysis correctly predicted that prednisolone could treat Crohn's disease, a condition for which it is a standard therapy.
Other matches were surprises. Butte's team chose to further investigate two such drug-disease combinations: an anti-ulcer medicine (cimetidine) that matched with lung cancer, and an anticonvulsant (topiramate) that matched with inflammatory bowel disease, which includes Crohn's disease.
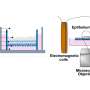
To confirm the cimetidine-lung cancer link, the team tested cimetidine on human lung cancer cells in lab dishes and implanted in mice. In both cases, the drug slowed the growth of the cancer cells compared to the control group (cells or mice) that had not received cimetidine.
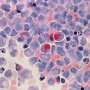
To test whether the anticonvulsant topiramate had an effect on inflammatory bowel diseases, the researchers administered the drug to rats that had bowel disease symptoms—diarrhea and inflammation, ulcers and microscopic damage in the colon. The drug decreased all of these symptoms, sometimes even better than prednisolone.
The work also has more fundamental value. The scientists noticed that diseases with similar molecular processes (for example, those that affect the immune system) clustered together in the analysis. So did drugs with similar effects (for example, those that slow cell division). The researchers believe that, by studying unexpected members of these clusters, they could learn more about how certain diseases progress and how some drugs work at the molecular level.
"This work is still at an early stage, but it is a promising proof of principle for a creative, fast and affordable approach to discovering new uses for drugs we already have in our therapeutic arsenal," Long said.
More information: Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, Sage J, Butte AJ. Discovery and Preclinical Validation of Drug Indications Using Compendia of Public Gene Expression Data. Sci Transl Med. 2011 Aug 17.
Dudley JT, Sirota M, Shenoy M, Pai R, Roedder S, Chiang AP, Morgan AA, Sarwal M, Pasricha PJ, Butte AJ. Computational Repositioning of the Anticonvulsant Topiramate for Inflammatory Bowel Disease. Sci Transl Med. 2011 Aug 17.