Research could lead to earlier diagnosis, treatment of mental diseases

A computer science and engineering associate professor and her doctoral student graduate are using a genetic computer network inference model that eventually could predict whether a person will suffer from bipolar disorder, schizophrenia or another mental illness.
The findings are detailed in the paper "Inference of SNP-Gene Regulatory Networks by Integrating Gene Expressions and Genetic Perturbations," which was published in the June edition of Biomed Research International. The principal investigators were Jean Gao, an associate professor of computer science and engineering, and Dong-Chul Kim, who recently earned his doctorate in computer science and engineering from UT Arlington.
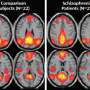
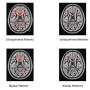
"We looked for the differences between our genetic computer network and the brain patterns of 130 patients from the University of Illinois," Gao said. "This work could lead to earlier diagnosis in the future and treatment for those patients suffering from bipolar disorder or schizophrenia. Early diagnosis allows doctors to provide timely treatments that may speed up aid to help affected patients."
The UT Arlington researchers teamed with Jiao Wang of the Beijing Genomics Institute at Wuhan, China; and Chunyu Liu, visiting associate professor at the University of Illinois Department of Psychiatry, on the project.
Gao said the findings also could lead to more individualized drug therapies for those patients in the early stages of mental illnesses.
"Our work will allow doctors to analyze a patient's genetic pattern and apply the appropriate levels of personalized therapy based on patient-specific data," Gao said.
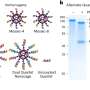
One key to the research is designing single nucleotide polymorphism or SNP networks, researchers said.
"SNPs are regulators of genes," said Kim, who joins the University of Texas-Pan American this fall as an assistant professor. "Those SNPs visualize how individual genes will act. It gives us more of a complete picture."
The paper is a culmination of four years of work.
Khosrow Behbehani, dean of the College of Engineering, said the research merges the power of computer science and engineering, psychology and genetics.
"This research holds a lot of promise in the area of genetic expression," Behbehani said. "If successful, it opens up the possibility of applying the method to other pathological conditions."
More information: Biomed Research International, www.hindawi.com/journals/bmri/2014/629697/