Structure mediating spread of antibiotic resistance identified
Scientists have identified the structure of a key component of the bacteria behind such diseases as whooping cough, peptic stomach ulcers and Legionnaires' disease. The research, funded by the Wellcome Trust and the Biotechnology and Biological Sciences Research Council (BBSRC), sheds light on how antibiotic resistance genes spread from one bacterium to another. The research may help scientists develop novel treatments for these diseases and novel ways to curtail the spread of antibiotic resistance.
Antibiotic resistance spreads when genetic material is exchanged between two bacteria, one of which has mutated to be resistant to the drugs. This exchange is facilitated by a multi-component device known as a type IV secretion system, which acts to transport antibiotic resistance genes from within one cell, through its membrane and into a neighbouring cell.
Type IV secretion systems also play an essential role in transporting toxins or proteins from within bacteria into the cells of the body, causing diseases. Examples of Gram-negative bacterial pathogens using such a device are Helicobacter pylori (which causes peptic ulcers), Legionella pneumophila (which causes Legionnaires' disease), and Bordetella pertussis (which causes whooping cough).
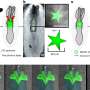
Now, in a paper published in the journal Science, scientists from the Institute of Structural and Molecular Biology (ISMB) at Birkbeck, University of London, and UCL (University College London) describe the structure of the core complex of a type IV secretion system, viewed using cryoelectron microscopy (a form of electron microscopy where the sample is studied at very low temperatures).
"Type IV secretion systems play key roles in secreting toxins which give certain bacteria their disease-causing properties and, importantly, are also directly involved in the spread of antibiotic resistance," says Professor Gabriel Waksman, Director of the ISMB and lead author of the study. "This is why they have become obvious targets in the vast effort required to fight infectious diseases caused by bacteria."
Gram-negative bacteria have a double membrane. At the core of the type IV secretion system is a double-walled chamber which spans the two membranes and opens at one side. Dr Waksman believes this chamber may offer a new pathway for targeting these bacteria.
"If we can inhibit the secretion systems that mediate transfer of antibiotic resistance genes from one bacterial pathogen to another, we could potentially prevent the spread of antibiotic resistance genes," he says. "For those pathogens that use type IV secretion system for secretion of toxins, the system can be targeted directly for inhibition. In both cases, this would have a considerable impact on public health."
Type IV secretion systems were first discovered in Agrobacterium tumefaciens, which uses the system to transfer tumour-inducing DNA into plants, causing "crown gall", which can be devastating to crops such as grape vines, sugar beet and rhubarb. However, crop scientists have been able to successfully exploit this transfer system as a way of introducing new genes into industrial crops, conferring herbicide-resistance and resistance to pathogens.
Source: Wellcome Trust