PET scanning probes reveal different cell function within the immune system
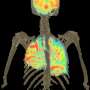
A commonly used probe for Positron Emission Tomography (PET) scanning and a new probe developed by researchers at UCLA reveal different functions in diverse cells of the immune system, providing a non-invasive and much clearer picture of an immune response in action.
The probes, the commonly used FDG that measures cellular glucose metabolism, and FAC, developed at UCLA and which measures the activity of a distinct biochemical pathway, work better when used in combination than either does alone. In addition to revealing the extent and cellular composition of an immune response, the probes also may be useful in evaluating therapies that target different cellular components of the immune system, said Dr. Owen Witte, a professor of microbiology, immunology and molecular genetics, a Howard Hughes Medical Institute investigator and senior author of the study.
"We demonstrated with this study that each probe targets different cells in the immune system with a high degree of specificity," said Witte, director of the UCLA Broad Stem Cell Research Center and a Jonsson Cancer Center researcher. "When cells are activated to do their job as an immune cell, the FDG probe is good at recognizing the subset of activated macrophages, while the FAC probe is good at recognizing the activated lymphocytes, as well as the macrophages. When tested sequentially, the combined information from the scans using the two probes gives you a better status of immune response."
The study, with lead author Evan Nair-Gill, a student in the campus' Medical Scientist Training Program, was conducted on mice bearing virally-induced sarcomas. The article appears today in the early online edition of the Journal of Clinical Investigation. Testing the probes in humans is the next step.
The scans provide clues to how the immune system works, for example, in response to cancer or auto-immune diseases such as rheumatoid arthritis, inflammatory bowel disease and multiple sclerosis, Witte said. They also could be used to see how therapies, such as vaccines and monoclonal antibodies meant to stimulate an immune response, are functioning within the body of a patient.
"This could give us another way to measure the efficacy of certain drugs," Witte said. "With some drugs, you could measure a change in the immune response within a week."
If the drugs are working, Witte said, doctors could stay the course. If they're not working or not working well enough, the therapy could be discontinued, sparing the patient a months-long exposure to an ineffective drug.
The next step will be testing the two probes in humans with a range of diseases, including cancer and auto-immune disorders, to confirm the work.
Witte and his colleagues licensed the FAC probe to Sofie Biosciences, which is owned in part by Witte and other UCLA faculty members. Researchers created the small molecule by slightly altering the molecular structure of one of the most commonly used chemotherapy drugs, gemcitabine. They then added a radiolabel so the cells that take in the probe can be seen during PET scanning.
The probe measures the activity of a fundamental cell biochemical pathway called the DNA salvage pathway, which acts as a recycling mechanism that helps with DNA replication and repair. All cells use this biochemical pathway to different degrees. But in lymphocytes and macrophages that are proliferating during an immune response, the pathway is activated to very high levels. Because of that, the probe accumulates at high levels in those cells, Witte said.