Molecular findings: Researchers identify region on gene that causes complex syndrome
(PhysOrg.com) -- Virginia Commonwealth University School of Medicine researchers have identified a gene that causes most of the features associated with brachydactyly mental retardation syndrome, a complex disorder involving developmental delays, autism spectrum disorder, sleep disturbance, skeletal anomalies, obesity and behavioral problems.
The findings could lead to the development of more cost-effective and efficient genetic testing that may help distinguish disorders with similar features.
In previous research, investigators have reported large deletions occurring on chromosome 2 as being associated with brachydactyly mental retardation syndrome. However, it has remained difficult to identify the precise region of the mutation, and therefore difficult to identify the specific gene that caused certain features of the disorder, until now.
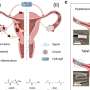
In a study published online in the August issue of the American Journal of Human Genetics, the VCU team reported that the cause of most of the features associated with the disorder may be due to a single gene called HDAC4, found within this deletion region. HDAC4 is known to regulate genes important in bone, muscle, neurological and cardiac development.
Through clinical and molecular analysis, the team examined six individuals with overlapping deletions involving chromosome 2, and were able to refine the critical region and reduce the number of candidate genes from more than 20 to a single gene - which was HDAC4. The team also reviewed scientific literature that provided information about other cases of this disorder.
Further, the study also linked two disorders together - Smith-Magenis syndrome and brachydactyly mental retardation syndrome - which were previously thought to be unrelated. Individuals with these two disorders are known to have very similar features such as developmental delays, obesity, sleep disturbances and behavioral problems. Smith-Magenis syndrome is a complex disorder associated with deletion or mutation of the RAI1 gene on chromosome 17.
“These findings are significant because of the difficulty in correctly identifying individuals for testing and proper diagnosis, and the lack of research on this group of complex syndromes associated with developmental delays and autism spectrum disorder,” said lead investigator, Sarah H. Elsea, Ph.D., associate professor in the VCU Departments of Pediatrics and Human and Molecular Genetics.
“We have made molecular connections between two disorders that were previously thought to be unrelated. By understanding the molecular etiology and the overlapping pathways between disorders such as these, we can focus efforts toward targeted therapeutics that may not only help one disorder, but several disorders with overlapping features due to defects in the same molecular and/or biochemical pathways,” she said.
According to Elsea, individuals with developmental delays, sleep disturbances, obesity and behavioral problems should have a genome-wide scan to detect changes in DNA copy number as a first test.
“This test will identify most cases of these disorders. If the genome-wide scan is negative, then direct DNA sequencing of specific genes should follow. These genome-wide tests are now becoming standard recommendations, but these two disorders illustrate the need for this process when diagnosing children and adults with developmental disabilities. It is a highly cost-effective process, with greater yield of confirmed diagnoses, which is important for the family,” Elsea said.