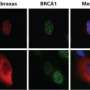
A new breast cancer susceptibility gene
Mutations in a gene called XRCC2 cause increased breast cancer risk, according to a study published today in the American Journal of Human Genetics. The study looked at families that have a history of the disease but do not have mutations in the currently known breast cancer susceptibility genes.
Sean Tavtigian, Ph.D., a Huntsman Cancer Institute (HCI) investigator and associate professor in the Department of Oncological Sciences at the University of Utah (U of U) is one of three co-principal investigators on the study, along with David Goldgar, Ph.D., professor in the Department of Dermatology at the U of U and an HCI investigator, and Melissa Southey, Ph.D., professor in the Department of Pathology at the University of Melbourne, Australia.
"We have added to the list of genes that harbour mutations causing breast cancer," said Tavtigian. "This knowledge will improve breast cancer diagnostics and add years to patients' lives. More important, relatives who have not been affected by the disease but carry the mutations will benefit even more. They can find out they are at risk before they have cancer and take action to reduce their risk or catch the cancer early."
XRCC2 may also provide a new target for chemotherapy. "A type of drug called a PARP inhibitor appears to kill tumor cells that have gene mutations in a particular DNA repair pathway. XRCC2 is in this pathway, as are BRCA1 and BRCA2. It's reasonably likely that a breast cancer patient who has a mutation in XRCC2 will respond well to treatment with PARP inhibitors," said Tavtigian.
According to Tavtigian, many breast cancer cases appear in families with a weak history of the disease. Only about 30 percent of the familial risk for breast cancer can be explained by a combination of mutations to and common sequence variation in the known breast cancer susceptibility genes. "So far most of the clinical diagnostic effort has been directed toward the very strong family history set of breast cancer cases and their close relatives," he said. "Our research looks at a population with a weaker family history, and as it turns out, a very rare gene mutation."
The researchers used a technology called exome capture massively parallel sequencing (exome sequencing), which shows the exact order of the nucleotides (the four building blocks of DNA) in all of the protein coding genes in the human genome. The ability of this technology to analyze the DNA of all of the genes in the genome in a single experiment, according to Tavtigian, makes it an amazingly powerful tool for genetic research. "We focused on the genes involved in a particular type of DNA repair, because most known breast cancer genes have been found there. That focused analysis allowed us to identify XRCC2 as a breast cancer susceptibility gene in individuals with a family history of breast cancer," says Tavtigian. "From the exome sequencing data, we found two different types of XRCC2 mutations that occur in breast cancer patients."
He explains that one type of mutation causes the gene to create an incomplete version of the protein. The resulting protein is usually dysfunctional. The other type occurs when a single amino acid in the protein is changed.
"It's a subtle change to the protein, but the resulting change in function could range anywhere from innocuous to even worse dysfunction than the incomplete protein causes," says Tavtigian. "Our sequence analyses suggest that we may have found the full spectrum of results in our study."
Further research is underway. "A worldwide effort has already been launched to figure out what fraction of breast cancer is due to mutations in this gene and how high the risk conferred by these mutations actually is," he says.